8DPJ
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001023030 | | Descriptor: | (2P)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-07-15 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8DIP
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001023030 | | Descriptor: | (2P)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5JLT
 
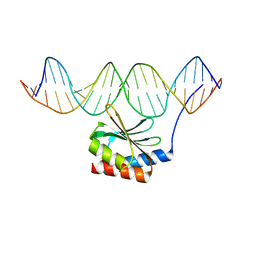 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
5JQ9
 
 | | Yersinia pestis DHPS with pterine-sulfa conjugate Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-N-(3,4-dimethyl-1,2-oxazol-5-yl)benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Wu, Y, White, S.W. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Pterin-sulfa conjugates as dihydropteroate synthase inhibitors and antibacterial agents.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CCY
 
 | | 2009 H1N1 PA endonuclease in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, THYMIDINE-5'-PHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-02 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CL0
 
 | | 2009 H1N1 PA endonuclease in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CGV
 
 | | 2009 H1N1 PA endonuclease in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5D42
 
 | | 2009 H1N1 PA endonuclease mutant F105S in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, THYMIDINE-5'-PHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DEB
 
 | | 2009 H1N1 PA endonuclease mutant E119D in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D2O
 
 | | 2009 H1N1 PA endonuclease mutant F105S | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-05 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D8U
 
 | | 2009 H1N1 PA endonuclease mutant E119D in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4G
 
 | | 2009 H1N1 PA endonuclease mutant F105S in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5D9J
 
 | | 2009 H1N1 PA endonuclease mutant F105S in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CZN
 
 | | 2009 H1N1 PA endonuclease mutant E119D | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DBS
 
 | | 2009 H1N1 PA endonuclease mutant E119D in complex with dTMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-21 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5DES
 
 | | 2009 H1N1 PA endonuclease domain | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6V53
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000985494 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000985494
To Be Published
|
|
6V56
 
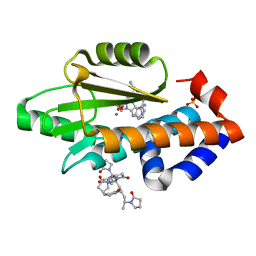 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000985494 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000985494
To Be Published
|
|
6VLL
 
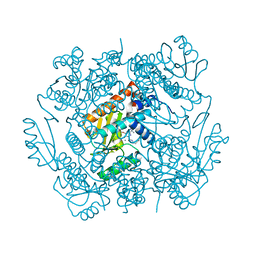 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant I38T in complex with SJ000986213 | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, SULFATE ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-24 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986213
To Be Published
|
|
7SNB
 
 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and ADP to 1.105 Angstrom resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Fatty Acid Kinase A, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and ADP to 1.105 Angstrom resolution
To Be Published
|
|
7UQ1
 
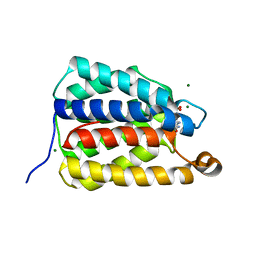 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and a single Mg ion at the dinuclear binding site | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fatty Acid Kinase A, GLYCEROL, ... | | Authors: | Cuypers, M.G, Subramanian, C, Seetharaman, J, Rock, C.O, White, S.W. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Domain architecture and catalysis of the Staphylococcus aureus fatty acid kinase.
J.Biol.Chem., 298, 2022
|
|
7RM7
 
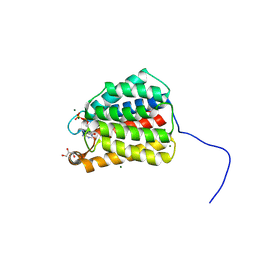 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.025 Angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fatty Acid Kinase A, GLYCEROL, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-07-26 | | Release date: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (1.025 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.025 Angstrom resolution
To Be Published
|
|
7RZK
 
 | | Co-soak crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.9 Angstrom resolution - SAD data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, Fatty Acid Kinase A, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-soak crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with ADP to 1.9 Angstrom resolution - SAD data
To Be Published
|
|
7SG3
 
 | | The X-ray crystal structure of the Staphylococcus aureus Fatty Acid Kinase B1 mutant A121I-A158L to 2.35 Angstrom resolution (Open form chain A, Palmitate bound) | | Descriptor: | Fatty Acid Kinase B1, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-04 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7SCL
 
 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase B1 (FakB1) mutant R173A in complex with Palmitate to 1.60 Angstrom resolution | | Descriptor: | Fatty Acid Kinase B1, GLYCEROL, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
