1I1S
 
 | | SOLUTION STRUCTURE OF THE TRANSCRIPTIONAL ACTIVATION DOMAIN OF THE BACTERIOPHAGE T4 PROTEIN MOTA | | Descriptor: | MOTA | | Authors: | Li, N, Zhang, W, White, S.W, Kriwacki, R.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-02-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA.
Biochemistry, 40, 2001
|
|
1KAF
 
 | | DNA Binding Domain Of The Phage T4 Transcription Factor MotA (AA105-211) | | Descriptor: | Transcription regulatory protein MOTA | | Authors: | Li, N, Sickmier, E.A, Zhang, R, Joachimiak, A, White, S.W. | | Deposit date: | 2001-11-01 | | Release date: | 2001-11-21 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the 'double wing' motif.
Mol.Microbiol., 43, 2002
|
|
1MMS
 
 | | Crystal structure of the ribosomal PROTEIN L11-RNA complex | | Descriptor: | 23S RIBOSOMAL RNA, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Wimberly, B.T, Guymon, R, Mccutcheon, J.P, White, S.W, Ramakrishnan, V. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A detailed view of a ribosomal active site: the structure of the L11-RNA complex.
Cell(Cambridge,Mass.), 97, 1999
|
|
7K0W
 
 | |
7KBC
 
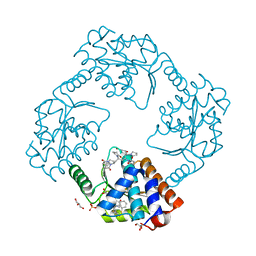 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D (construct with truncated loop 51-72) in complex with baloxavir acid | | Descriptor: | Baloxavir acid, GLYCEROL, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Kumar, G, Slavish, J, White, S.W. | | Deposit date: | 2020-10-02 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7KAF
 
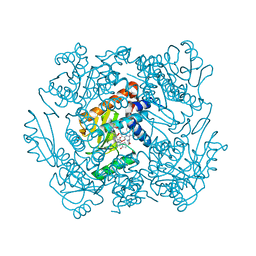 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T (construct with truncated loop 51-72) in complex with baloxavir acid | | Descriptor: | Baloxavir acid, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Slavish, J, White, S.W. | | Deposit date: | 2020-09-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7KL3
 
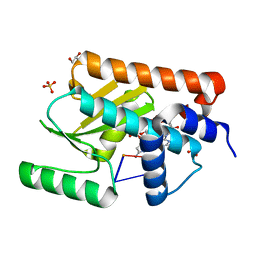 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D bound to RNA oligomer AG*CAUC (*uncleaveable bond, -UC disordered) | | Descriptor: | 5'-O-sulfocytidine, ADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cuypers, M.G, Kumar, G, White, S.W. | | Deposit date: | 2020-10-28 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7K87
 
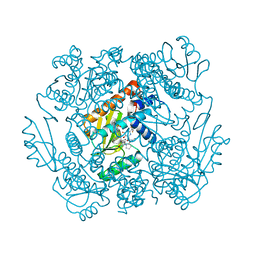 | | The crystal structure of the 2009 H1N1 PA endonuclease in complex with SJ000986436 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7LW6
 
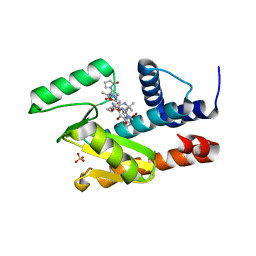 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with Raltegravir | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, White, S.W, Rankovik, Z. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
1BM9
 
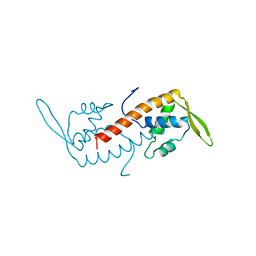 | |
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
487D
 
 | |
1QD7
 
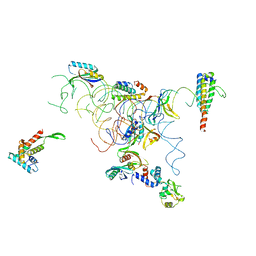 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
5W7U
 
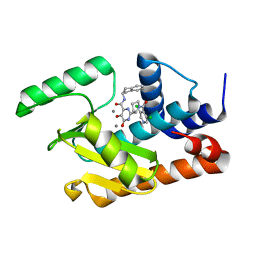 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 8f (SRI-29928) | | Descriptor: | 2-[(2S)-1-(3,5-dichloropyridine-4-carbonyl)pyrrolidin-2-yl]-N-(2,3-dihydro-1H-inden-2-yl)-5-hydroxy-6-oxo-1,6-dihydropy rimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W9G
 
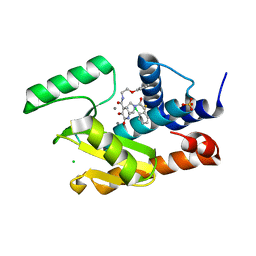 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9k (SRI-30023) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4 -carboxamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W73
 
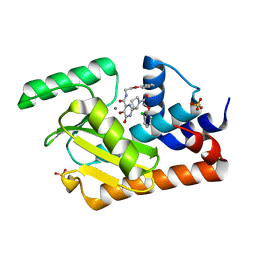 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9f (SRI-29835) | | Descriptor: | 2-{(2S)-1-[(2-chlorophenoxy)acetyl]pyrrolidin-2-yl}-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W92
 
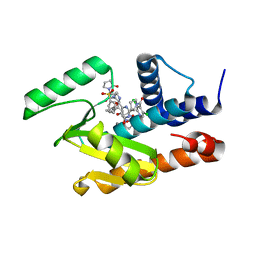 | | Crystal structure of the influenza virus PA endonuclease in complex with an inhibitor - SRI-30049 | | Descriptor: | 1-[(3R,5S,7R)-1,5,7,9-tetrakis(2-oxopyrrolidin-1-yl)nonan-3-yl]-1,3-dihydro-2H-pyrrol-2-one, 2-[(2S)-1-(3,5-dichloropyridine-4-carbonyl)pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-[2-(phenylsulfonyl)ethyl]-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
1FKA
 
 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
4X9X
 
 | | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid Binding Protein Family | | Descriptor: | 1,2-ETHANEDIOL, DegV domain-containing protein MW1315, OLEIC ACID | | Authors: | Broussard, T.C, Miller, D.J, Jackson, P, Nourse, A, Rock, C.O. | | Deposit date: | 2014-12-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Biochemical Roles for Conserved Residues in the Bacterial Fatty Acid-binding Protein Family.
J.Biol.Chem., 291, 2016
|
|
2EWS
 
 | |
1RSS
 
 | |
3IO5
 
 | |
6NXD
 
 | |
2AQ7
 
 | | Structure-activity relationships at the 5-posiiton of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherichia coli | | Descriptor: | (5R)-4-HYDROXY-3,5-DIMETHYL-5-[(1E,3E)-2-METHYLPENTA-1,3-DIENYL]THIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
2AQB
 
 | | Structure-activity relationships at the 5-position of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherchia coli | | Descriptor: | (5R)-5-[(1E)-BUTA-1,3-DIENYL]-4-HYDROXY-3,5-DIMETHYLTHIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
