3S53
 
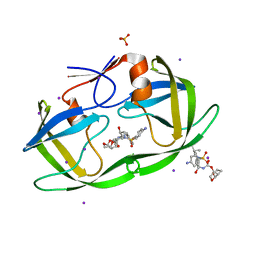 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P212121 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, IODIDE ION, PHOSPHATE ION, ... | | 著者 | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | 登録日 | 2011-05-20 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S43
 
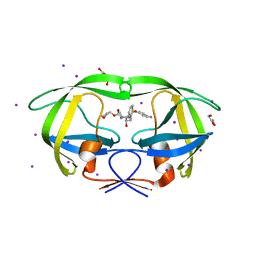 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug amprenavir | | 分子名称: | GLYCEROL, IODIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Tie, Y.-F, Weber, I.T. | | 登録日 | 2011-05-18 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3SM8
 
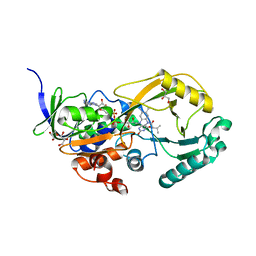 | |
3ST5
 
 | | Crystal structure of wild-type HIV-1 protease with C3-Substituted Hexahydrocyclopentafuranyl Urethane as P2-Ligand, GRL-0489A | | 分子名称: | (3R,3aR,5R,6aR)-3-hydroxyhexahydro-2H-cyclopenta[b]furan-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2011-07-08 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Design of HIV-1 Protease Inhibitors with C3-Substituted Hexahydrocyclopentafuranyl Urethanes as P2-Ligands: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Crystal Structure.
J.Med.Chem., 54, 2011
|
|
8HVP
 
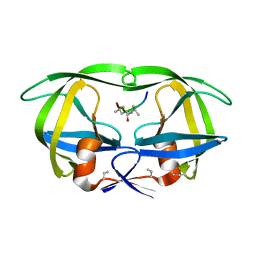 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | 分子名称: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | 著者 | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | 登録日 | 1990-10-26 | | 公開日 | 1993-10-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
6BKA
 
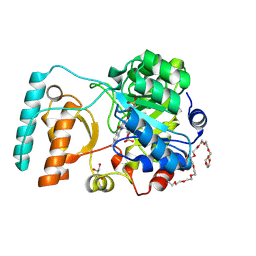 | | Crystal Structure of Nitronate Monooxygenase from Cyberlindnera saturnus | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Agniswamy, J, Fang, Y.-F, Weber, I.T. | | 登録日 | 2017-11-08 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of yeast nitronate monooxygenase from Cyberlindnera saturnus.
Proteins, 86, 2018
|
|
7HVP
 
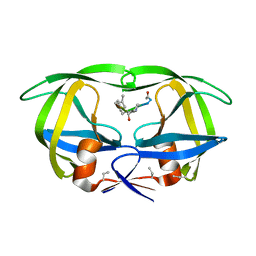 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | 分子名称: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | 著者 | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | 登録日 | 1990-09-13 | | 公開日 | 1993-07-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
6E2A
 
 | | Crystal structure of NADH:quinone reductase PA1024 from Pseudomonas aeruginosa PAO1 in complex with NAD+ | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Reis, R.A.G, Ball, J, Agniswamy, J, Weber, I, Gadda, G. | | 登録日 | 2018-07-10 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Steric hindrance controls pyridine nucleotide specificity of a flavin-dependent NADH:quinone oxidoreductase.
Protein Sci., 28, 2019
|
|
2REB
 
 | |
2QLJ
 
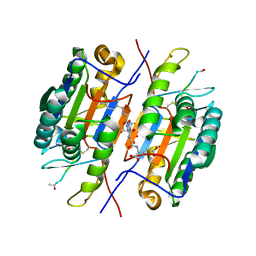 | | Crystal Structure of Caspase-7 with Inhibitor AC-WEHD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-WEHD-CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL5
 
 | | Crystal Structure of caspase-7 with inhibitor AC-DMQD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, inhibitor, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL9
 
 | | Crystal Structure of Caspase-7 with inhibitor AC-DQMD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-DQMD-CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QLF
 
 | |
2QLB
 
 | | Crystal Structure of caspase-7 with inhibitor AC-ESMD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-ESMD-CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL7
 
 | | Crystal Structure of Caspase-7 with inhibitor AC-IEPD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-IEPD_CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
5T8H
 
 | |
6PLD
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with 6-OH-FAD - Green fraction | | 分子名称: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | 著者 | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | 登録日 | 2019-06-30 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6P9D
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | 著者 | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | 登録日 | 2019-06-10 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.329 Å) | | 主引用文献 | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
3HVP
 
 | |
3TKW
 
 | | Crystal structure of HIV protease model precursor/Darunavir complex | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3TL9
 
 | | crystal structure of HIV protease model precursor/Saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3UHL
 
 | |
1REA
 
 | |
2G69
 
 | |
3IBF
 
 | | Crystal structure of unliganded caspase-7 | | 分子名称: | Caspase-7 | | 著者 | Agniswamy, J. | | 登録日 | 2009-07-15 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Conformational similarity in the activation of caspase-3 and -7 revealed by the unliganded and inhibited structures of caspase-7.
Apoptosis, 14, 2009
|
|
