6BNF
 
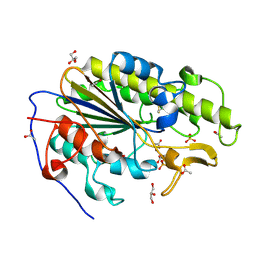 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, mono-zinc complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
6AOO
 
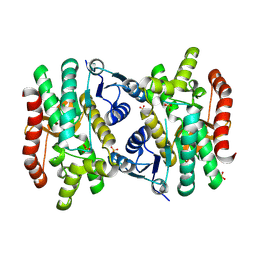 | | 2.15 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae | | Descriptor: | Malate dehydrogenase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae.
To Be Published
|
|
6B3P
 
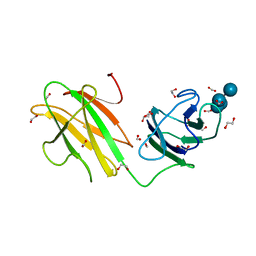 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
6AZI
 
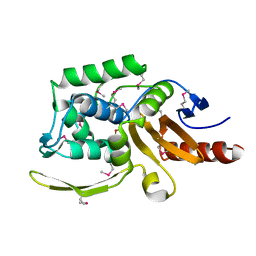 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
6AON
 
 | | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD | | Descriptor: | CALCIUM ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom Resolution Crystal Structure of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase from Bordetella pertussis in Complex with FAD
To Be Published
|
|
6AZ5
 
 | |
6BAL
 
 | | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CHLORIDE ION, Malate dehydrogenase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate
To Be Published
|
|
6B15
 
 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K | | Descriptor: | 1,2-ETHANEDIOL, Amy13K | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-16 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
6BND
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant mono-zinc and phosphoethanolamine complex | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6BNE
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, phosphate-bound complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
4MH6
 
 | | 2.8 Angstrom Crystal Structure of Type III Secretion Protein YscO from Vibrio parahaemolyticus | | Descriptor: | PHOSPHATE ION, Putative type III secretion protein YscO | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Winsor, J, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 Angstrom Crystal Structure of Type III Secretion Protein YscO from Vibrio parahaemolyticus.
TO BE PUBLISHED
|
|
4MHB
 
 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
4MJZ
 
 | | 2.75 Angstrom Resolution Crystal Structure of Putative Orotidine-monophosphate-decarboxylase from Toxoplasma gondii. | | Descriptor: | DI(HYDROXYETHYL)ETHER, NITRATE ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Minasov, G, Wawrzak, Z, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Putative Orotidine-monophosphate-decarboxylase from Toxoplasma gondii.
TO BE PUBLISHED
|
|
4NVR
 
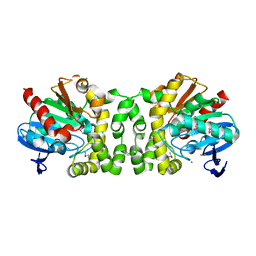 | | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Stam, J, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica.
TO BE PUBLISHED
|
|
4OMV
 
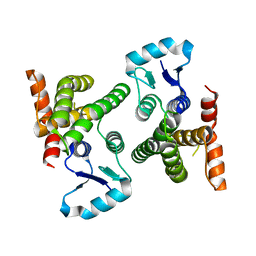 | | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4 | | Descriptor: | Macrophage growth locus, subunit A | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4
TO BE PUBLISHED
|
|
4OVD
 
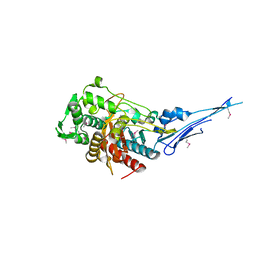 | | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469 | | Descriptor: | CALCIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Babnigg, G, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469
To be Published
|
|
5E31
 
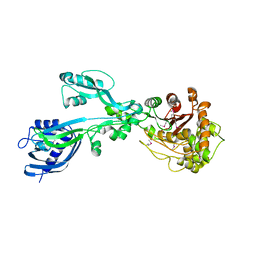 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
4N3O
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni. | | Descriptor: | CALCIUM ION, Putative D-glycero-D-manno-heptose 7-phosphate kinase | | Authors: | Minasov, G, Wawrzak, Z, Gordon, E, Onopriyenko, O, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-07 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.
TO BE PUBLISHED
|
|
5DVY
 
 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5EQ4
 
 | | Crystal structure of the SrpA adhesin R347E mutant from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ2
 
 | | Crystal Structure of the SrpA Adhesin from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
4OFX
 
 | | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii | | Descriptor: | Cystathionine beta-synthase, SODIUM ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii
To be Published
|
|
5EQ3
 
 | | Crystal structure of the SrpA adhesin from Streptococcus sanguinis with a sialyl galactose disaccharide bound | | Descriptor: | ACETATE ION, CALCIUM ION, N-glycolyl-alpha-neuraminic acid-(2-3)-methyl beta-D-galactopyranoside, ... | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
4MFG
 
 | | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.
TO BE PUBLISHED
|
|
