3CWS
 
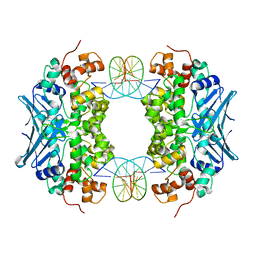 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxyinosine:Thymine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FI)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DTP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CW7
 
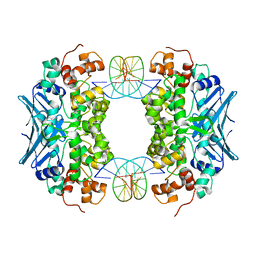 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CWT
 
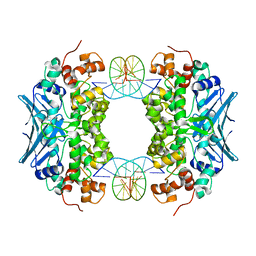 | | Crystal Structure of an AlkA Host/Guest Complex 2'-fluoro-2'-deoxyinosine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(2FI)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CVS
 
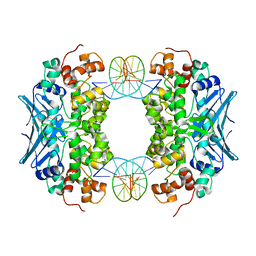 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CWA
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*(8OG)P*DAP*DCP*DAP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3D4V
 
 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
1GYX
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1JQ0
 
 | | Mutation that destabilize the gp41 core: determinants for stabilizing the SIV/CPmac envelope glycoprotein complex. Mutant structure. | | Descriptor: | gp41 envelope protein | | Authors: | Liu, J, Wang, S, LaBranche, C.C, Hoxie, J.A, Lu, M. | | Deposit date: | 2001-08-03 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex.
J.Biol.Chem., 277, 2002
|
|
1JPX
 
 | | Mutation that destabilize the gp41 core: determinants for stabilizing the SIV/CPmac envelope glycoprotein complex. Wild type. | | Descriptor: | gp41 envelope protein | | Authors: | Liu, J, Wang, S, LaBranche, C.C, Hoxie, J.A, Lu, M. | | Deposit date: | 2001-08-03 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex.
J.Biol.Chem., 277, 2002
|
|
3QYC
 
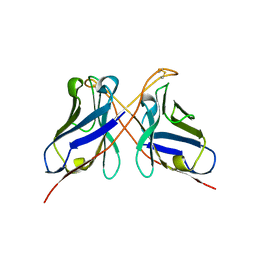 | | Structure of a dimeric anti-HER2 single domain antibody | | Descriptor: | VH domain of IgG molecule | | Authors: | Baral, T.N, Chao, S, Li, S, Tanha, J, Arbabai, M, Wang, S, Zhang, J. | | Deposit date: | 2011-03-03 | | Release date: | 2012-02-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Human Single Domain Antibody Dimer Formed through V(H)-V(H) Non-Covalent Interactions.
Plos One, 7, 2012
|
|
4QXA
 
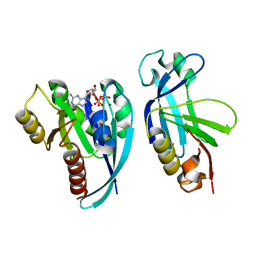 | | Crystal structure of the Rab9A-RUTBC2 RBD complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-9A, ... | | Authors: | Zhang, Z, Wang, S, Ding, J. | | Deposit date: | 2014-07-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Rab9A-RUTBC2 RBD complex reveals the molecular basis for the binding specificity of Rab9A with RUTBC2.
Structure, 22, 2014
|
|
1OR7
 
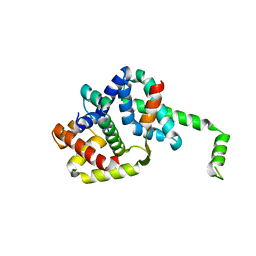 | | Crystal Structure of Escherichia coli sigmaE with the Cytoplasmic Domain of its Anti-sigma RseA | | Descriptor: | RNA polymerase sigma-E factor, Sigma-E factor negative regulatory protein | | Authors: | Campbell, E.A, Tupy, J.L, Gruber, T.M, Wang, S, Sharp, M.M, Gross, C.A, Darst, S.A. | | Deposit date: | 2003-03-12 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA.
Mol.Cell, 11, 2003
|
|
7XJF
 
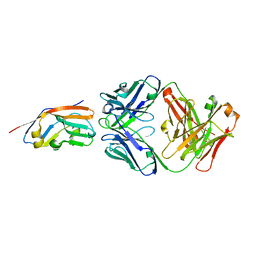 | | Crystal structure of 6MW3211 Fab in complex with CD47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2022-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment.
Theranostics, 13, 2023
|
|
1PXI
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,5-Dichloro-thiophen-3-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,5-DICHLOROTHIEN-3-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1PW2
 
 | | APO STRUCTURE OF HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-06-30 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the
activation loop.
Structure, 11, 2003
|
|
1PXJ
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
3CM1
 
 | |
1PXK
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)pyrimidin-2-yl]-N'-hydroxyiminoformamide | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-YL]-N'-HYDROXYIMIDOFORMAMIDE | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1PXL
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-(4-trifluoromethyl-phenyl)-amine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)-N-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1RHT
 
 | | 24-MER RNA HAIRPIN COAT PROTEIN BINDING SITE FOR BACTERIOPHAGE R17 (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RNA (5'-R(P*GP*GP*GP*AP*CP*UP*GP*AP*CP*GP*AP*UP*CP*AP*CP*GP*CP*AP*GP*UP*CP*UP*AP*U)-3') | | Authors: | Borer, P.N, Lin, Y, Wang, S, Roggenbuck, M.W, Gott, J.M, Uhlenbeck, O.C, Pelczer, I. | | Deposit date: | 1995-03-03 | | Release date: | 1995-06-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Biochemistry, 34, 1995
|
|
3OH6
 
 | | AlkA Undamaged DNA Complex: Interrogation of a C:G base pair | | Descriptor: | 5'-D(*GP*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*CP*AP*TP*TP*CP*AP*TP*GP*TP*C)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
3OH9
 
 | | AlkA Undamaged DNA Complex: Interrogation of a T:A base pair | | Descriptor: | 5'-D(*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*TP*TP*CP*AP*TP*GP*T)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
8GLT
 
 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of energy transfer proteins housing excitonically coupled chlorophyll special pairs
To Be Published
|
|
3OGD
 
 | | AlkA Undamaged DNA Complex: Interrogation of a G*:C base pair | | Descriptor: | 5'-D(*CP*AP*(BRU)P*GP*AP*CP*(BRU)P*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*CP*AP*TP*G)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
3R0J
 
 | | Structure of PhoP from Mycobacterium tuberculosis | | Descriptor: | POSSIBLE TWO COMPONENT SYSTEM RESPONSE TRANSCRIPTIONAL POSITIVE REGULATOR PHOP, R-1,2-PROPANEDIOL, SULFATE ION | | Authors: | Menon, S, Wang, S. | | Deposit date: | 2011-03-08 | | Release date: | 2011-07-13 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Response Regulator PhoP from Mycobacterium tuberculosis Reveals a Dimer through the Receiver Domain.
Biochemistry, 50, 2011
|
|
