8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wei, H, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Wei, H, Guo, M, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
1Q2F
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | 分子名称: | PNC27 | | 著者 | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | 登録日 | 2003-07-24 | | 公開日 | 2004-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
5YIJ
 
 | | Structure of a Legionella effector with substrates | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | 著者 | Feng, Y, Mu, Y, Wang, H. | | 登録日 | 2017-10-05 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
1LP9
 
 | | Xenoreactive complex AHIII 12.2 TCR bound to p1049/HLA-A2.1 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | 著者 | Buslepp, J, Wang, H, Biddison, W.E, Appella, E, Collins, E.J. | | 登録日 | 2002-05-07 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection.
Immunity, 19, 2003
|
|
1MPV
 
 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | 分子名称: | BLyS Receptor 3 | | 著者 | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | 登録日 | 2002-09-12 | | 公開日 | 2002-10-30 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
3TDL
 
 | | Structure of human serum albumin in complex with DAUDA | | 分子名称: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | 著者 | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | 登録日 | 2011-08-11 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
4H58
 
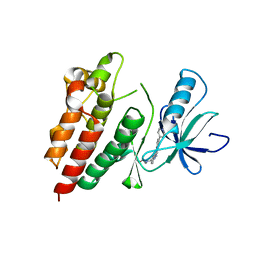 | | BRAF in complex with compound 3 | | 分子名称: | CHLORIDE ION, N-(4-{[(2-methoxyethyl)amino]methyl}phenyl)-6-(pyridin-4-yl)quinazolin-2-amine, Serine/threonine-protein kinase B-raf | | 著者 | Vasbinder, M, Aquila, B, Augustin, M, Chueng, T, Cook, D, Drew, L, Fauber, B, Glossop, S, Godin, R, Grondine, M, Hennessy, E, Johannes, J, Lee, S, Lyne, P, Moertl, M, Omer, C, Palakurthi, S, Pontz, T, Read, J, Sha, L, Shen, M, Steinbacher, S, Wang, H, Wu, A, Ye, M, Bagal, B. | | 登録日 | 2012-09-18 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf(V600E) Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
2K8U
 
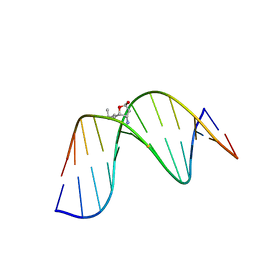 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6S,8R,11S)-configuration matched with dC | | 分子名称: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | 著者 | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | 登録日 | 2008-09-23 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
2KAS
 
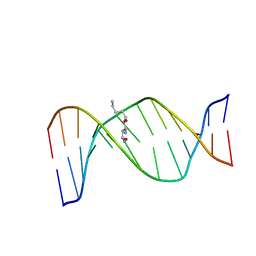 | | HNE-dG adduct mismatched with dA in basic solution | | 分子名称: | (4S)-nonane-1,4-diol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DAP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | 著者 | Huang, H, Wang, H, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | 登録日 | 2008-11-13 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA
Chem.Res.Toxicol., 22, 2009
|
|
2K8T
 
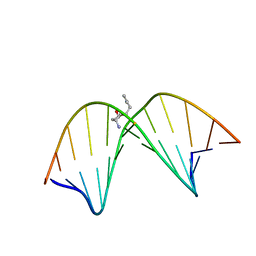 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6R,8S,11R)-configuration opposite dC | | 分子名称: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | 著者 | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | 登録日 | 2008-09-23 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
2KAR
 
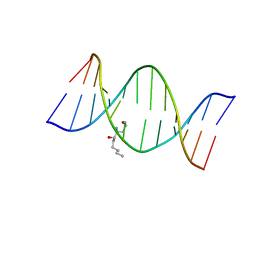 | | HNE-dG adduct mismatched with dA in acidic solution | | 分子名称: | (4S)-nonane-1,4-diol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DAP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | 著者 | Huang, H, Wang, H, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | 登録日 | 2008-11-13 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA
Chem.Res.Toxicol., 22, 2009
|
|
1VLZ
 
 | |
8HXX
 
 | |
8HXY
 
 | |
8HY0
 
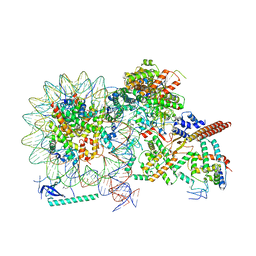 | |
8HXZ
 
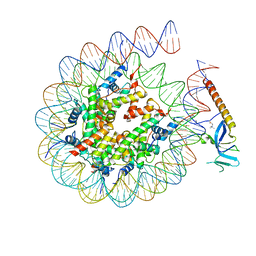 | |
4PY9
 
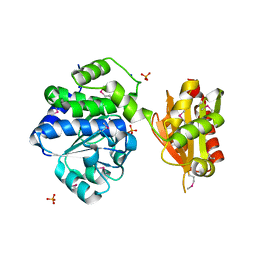 | | Crystal structure of an exopolyphosphatase-related protein from Bacteroides Fragilis. Northeast Structural Genomics target BFR192 | | 分子名称: | PHOSPHATE ION, Putative exopolyphosphatase-related protein, SODIUM ION | | 著者 | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-03-26 | | 公開日 | 2014-06-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of an exopolyphosphatase-related protein from Bacteroides Fragilis. Northeast Structural Genomics target BFR192
To be Published
|
|
2LBI
 
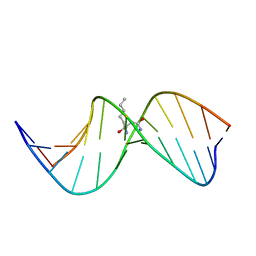 | | N2-dG:N2-dG interstrand cross-link induced by trans-4-hydroxynonenal | | 分子名称: | (4S)-nonane-1,4-diol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*CP*GP*CP*TP*AP*GP*C)-3') | | 著者 | Huang, H, Kozekov, I.D, Wang, H, Kozekova, A, Rizzo, C.J, Stone, M.P. | | 登録日 | 2011-03-31 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Formation of a N2-dG:N2-dG carbinolamine DNA cross-link by the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N2-dG adduct.
J.Am.Chem.Soc., 133, 2011
|
|
3B55
 
 | | Crystal structure of the Q81BN2_BACCR protein from Bacillus cereus. NESG target BcR135 | | 分子名称: | CALCIUM ION, Succinoglycan biosynthesis protein | | 著者 | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Wang, H, Cunningham, K, Owens, L, Maglaqui, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-10-25 | | 公開日 | 2007-11-06 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the Q81BN2_BACCR protein from Bacillus cereus.
To be Published
|
|
2WOC
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | 分子名称: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | 登録日 | 2009-07-23 | | 公開日 | 2009-08-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOE
 
 | | Crystal Structure of the D97N variant of dinitrogenase reductase- activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribose | | 分子名称: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | 登録日 | 2009-07-23 | | 公開日 | 2009-08-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOD
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP- ribsoyllysine | | 分子名称: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | 登録日 | 2009-07-23 | | 公開日 | 2009-08-11 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3CEW
 
 | | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205 | | 分子名称: | Uncharacterized cupin protein, ZINC ION | | 著者 | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-02-29 | | 公開日 | 2008-03-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205.
To be Published
|
|
3CNW
 
 | | Three-dimensional structure of the protein XoxI (Q81AY6) from Bacillus cereus. Northeast Structural Genomics Consortium target BcR196. | | 分子名称: | Protein XoxI | | 著者 | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Ciccosanti, C, Mao, L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-03-26 | | 公開日 | 2008-04-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Three-dimensional structure of the protein XoxI (Q81AY6) from Bacillus cereus. Northeast Structural Genomics Consortium target BcR196.
To Be Published
|
|
