1CNP
 
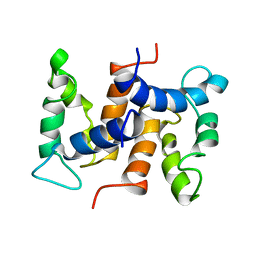 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
1DSI
 
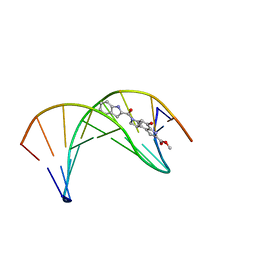 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
1EIS
 
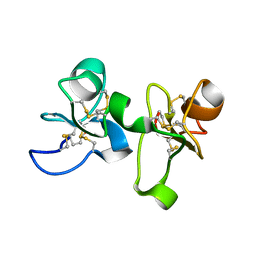 | | UDA UNCOMPLEXED FORM. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | PROTEIN (AGGLUTININ ISOLECTIN VI/AGGLUTININ ISOLECTIN V) | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-02-28 | | Release date: | 2000-06-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1ENM
 
 | | UDA TRISACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-06-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1EN2
 
 | | UDA TETRASACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | Authors: | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | Deposit date: | 2000-03-20 | | Release date: | 2000-06-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
1PFL
 
 | | REFINED SOLUTION STRUCTURE OF HUMAN PROFILIN I | | Descriptor: | PROFILIN I | | Authors: | Metzler, W.J, Farmer II, B.T, Constantine, K.L, Friedrichs, M.S, Lavoie, T, Mueller, L. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of human profilin I.
Protein Sci., 4, 1995
|
|
1FNZ
 
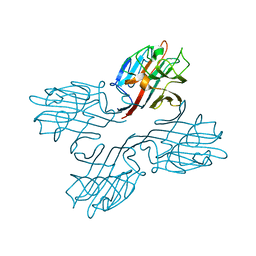 | | A bark lectin from robinia pseudoacacia in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, BARK AGGLUTININ I, POLYPEPTIDE A, ... | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2000-08-24 | | Release date: | 2001-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine
Proteins, 44, 2001
|
|
1FNY
 
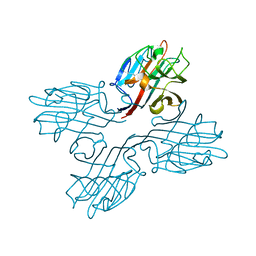 | | LEGUME LECTIN OF THE BARK OF ROBINIA PSEUDOACACIA. | | Descriptor: | BARK AGGLUTININ I,POLYPEPTIDE A, CALCIUM ION | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2000-08-24 | | Release date: | 2001-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine.
Proteins, 44, 2001
|
|
8PDG
 
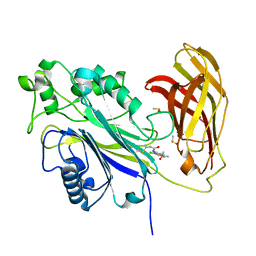 | | The phosphatase and C2 domains of SHIP1 with covalent Z2738285202 | | Descriptor: | DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1, ~{N}-(8-chloranylquinolin-2-yl)propanamide | | Authors: | Bradshaw, W.J, Moreira, T, Scacioc, A, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
8PDH
 
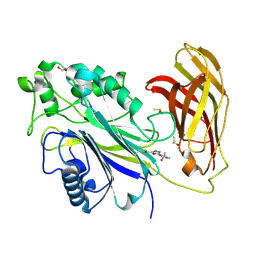 | | The phosphatase and C2 domains of SHIP1 with covalent Z1742148362 | | Descriptor: | (5-phenyl-1,3,4-oxadiazol-2-yl)methanimine, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
8PDJ
 
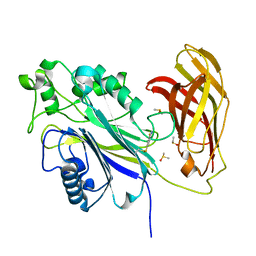 | | The phosphatase and C2 domains of SHIP1 with covalent Z56948267 | | Descriptor: | 4-azanyl-3-fluoranyl-benzenethiol, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
8PDI
 
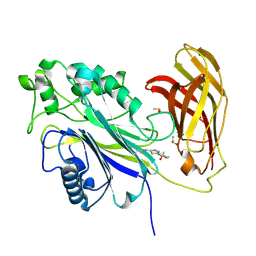 | | The phosphatase and C2 domains of SHIP1 with covalent Z1763271112 | | Descriptor: | (5-phenyl-1,3,4-thiadiazol-2-yl)methanimine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
8SDK
 
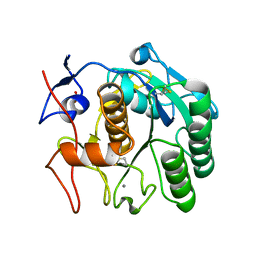 | | The MicroED structure of proteinase K crystallized by suspended drop crystallization | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Gillman, C, Nicolas, W.J, Martynowycz, M.W, Gonen, T. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Design and implementation of suspended drop crystallization.
Iucrj, 10, 2023
|
|
8SB6
 
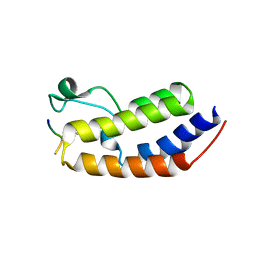 | | Structure of human BRD2-BD1 bound to a histone H4 acetyl-methyllysine peptide | | Descriptor: | Bromodomain containing 2, Histone H4 | | Authors: | Connor, L.J, Ekundayo, B.E, Lu-Culligan, W.J, Simon, M.D, Bleichert, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622, 2023
|
|
8S9W
 
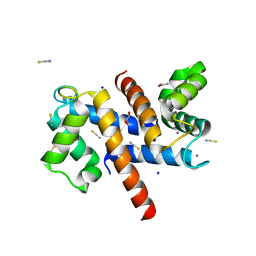 | | Murine S100A7/S100A15 in presence of calcium | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Harrison, S.A, Naretto, A, Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-03-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Comparative analysis of the physical properties of murine and human S100A7: Insight into why zinc piracy is mediated by human but not murine S100A7.
J.Biol.Chem., 299, 2023
|
|
3GAE
 
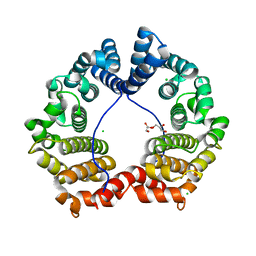 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GM7
 
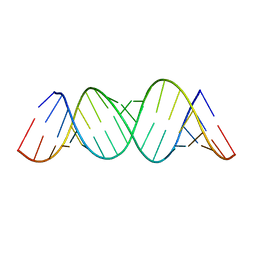 | |
3GT2
 
 | | Crystal Structure of the P60 Domain from M. avium paratuberculosis Antigen MAP1272c | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, SULFATE ION | | Authors: | Ramyar, K.X, Lingle, C.K, McWhorter, W.J, Bouyain, S, Bannantine, J.P, Geisbrecht, B.V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Two P60-Family Antigens from Mycobacterium Avium Paratuberculosis
To be Published
|
|
3GLP
 
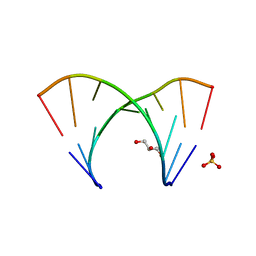 | | 1.23 A resolution X-ray structure of (GCUGCUGC)2 | | Descriptor: | 5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3', GLYCEROL, SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into CUG repeats containing the 'stretched U-U wobble': implications for myotonic dystrophy.
Nucleic Acids Res., 37, 2009
|
|
3H44
 
 | |
3HGZ
 
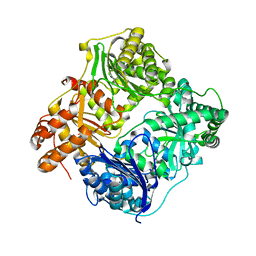 | | Crystal structure of human insulin-degrading enzyme in complex with amylin | | Descriptor: | Insulin-degrading enzyme, Islet amyloid polypeptide, ZINC ION | | Authors: | Guo, Q, Bian, Y, Tang, W.J. | | Deposit date: | 2009-05-14 | | Release date: | 2009-12-08 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
3HQB
 
 | |
3HQA
 
 | |
3I86
 
 | | Crystal structure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 | | Descriptor: | ISOPROPYL ALCOHOL, Putative uncharacterized protein | | Authors: | Ramyar, K.X, Lingle, C.K, McWhorter, W.J, Bouyain, S, Bannantine, J.P, Geisbrecht, B.V. | | Deposit date: | 2009-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atypical structural features of two P60 family members from Mycobacterium avium subspecies paratuberculosis
To be Published
|
|
3IYS
 
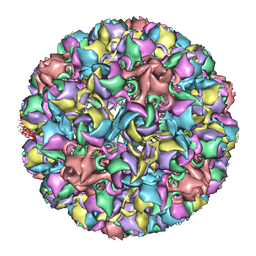 | | Homology model of avian polyomavirus asymmetric unit | | Descriptor: | Major capsid protein VP1 | | Authors: | Shen, P.S, Enderlein, D, Nelson, C.D.S, Carter, W.S, Kawano, M, Xing, L, Swenson, R.D, Olson, N.H, Baker, T.S, Cheng, R.H, Atwood, W.J, Johne, R, Belnap, D.M. | | Deposit date: | 2010-04-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | The structure of avian polyomavirus reveals variably sized capsids, non-conserved inter-capsomere interactions, and a possible location of the minor capsid protein VP4.
Virology, 411, 2011
|
|
