3VZQ
 
 | |
3VZP
 
 | | Crystal structure of PhaB from Ralstonia eutropha | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Acetoacetyl-CoA reductase, GLYCEROL, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3VZS
 
 | | Crystal structure of PhaB from Ralstonia eutropha in complex with Acetoacetyl-CoA and NADP | | Descriptor: | ACETOACETYL-COENZYME A, Acetoacetyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
8KG3
 
 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
6FEK
 
 | |
8AMQ
 
 | | Crystal structure of the complex CYP143-FdxE from M. tuberculosis | | Descriptor: | FE3-S4 CLUSTER, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Smolskaya, S, Marin, E, Kapranov, I, Kovalev, K, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMP
 
 | | Crystal structure of M.tuberculosis ferredoxin Fdx | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, Possible ferredoxin | | Authors: | Bukhdruker, S, Kavaleuski, A, Marin, E, Kapranov, I, Mishin, A, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8AMO
 
 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
4LNR
 
 | | The structure of HLA-B*35:01 in complex with the peptide (RPQVPLRPMTY) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Cheng, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-12 | | Release date: | 2014-07-23 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex.
J.Biol.Chem., 289, 2014
|
|
5I20
 
 | | Crystal structure of protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | Authors: | Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
7A9A
 
 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
7XTX
 
 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
2MG2
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N.L, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
2MG3
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
2NCT
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2NCS
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2MG1
 
 | | NMR assignment and structure of a peptide derived from the trans-membrane region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N.L, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
8J6N
 
 | | Crystal structure of Cystathionine gamma-lyase in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, Cystathionine gamma-lyase, GLYCEROL, ... | | Authors: | Hibi, R, Toma-Fukai, S, Shimizu, T, Hanaoka, K. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a cystathionine gamma-lyase (CSE) selective inhibitor targeting active-site pyridoxal 5'-phosphate (PLP) via Schiff base formation.
Sci Rep, 13, 2023
|
|
8IMR
 
 | | Structure of ligand-free human macrophage migration inhibitory factor | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
1WV3
 
 | |
7FB7
 
 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
5EW1
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5EW2
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5GJ3
 
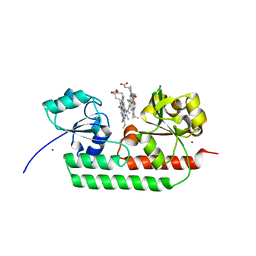 | | Periplasmic heme-binding protein RhuT from Roseiflexus sp. RS-1 in two-heme bound form (holo-2) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Periplasmic binding protein, ZINC ION | | Authors: | Rahman, M.M, Naoe, Y, Nakamura, N, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
5GIZ
 
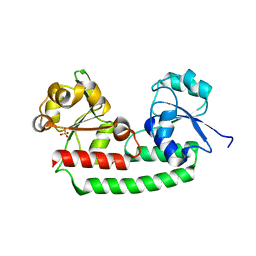 | | Periplasmic heme-binding protein BhuT in apo form | | Descriptor: | CHLORIDE ION, Putative hemin transport system, substrate-binding protein, ... | | Authors: | Nakamura, N, Naoe, Y, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
