3EFK
 
 | | Structure of c-Met with pyrimidone inhibitor 50 | | 分子名称: | 5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-2-[(4-fluorophenyl)amino]-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | 著者 | Bellon, S.F, D'Angelo, N, Whittington, D, Dussault, I. | | 登録日 | 2008-09-09 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
3EFJ
 
 | | Structure of c-Met with pyrimidone inhibitor 7 | | 分子名称: | 2-benzyl-5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | 著者 | D'Angelo, N, Bellon, S, Whittington, D. | | 登録日 | 2008-09-09 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
8T5C
 
 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-06-13 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
3S0Z
 
 | | Crystal structure of New Delhi Metallo-beta-lactamase (NDM-1) | | 分子名称: | Metallo-beta-lactamase, ZINC ION | | 著者 | Guo, Y, Wang, J, Niu, G.J, Shui, W.Q, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | 登録日 | 2011-05-13 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A structural view of the antibiotic degradation enzyme NDM-1 from a superbug.
Protein Cell, 2011
|
|
6OZB
 
 | | Crystal structure of the phycoerythrobilin-bound GAF domain from a cyanobacterial phytochrome | | 分子名称: | PHYCOERYTHROBILIN, Two-component sensor histidine kinase | | 著者 | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | 登録日 | 2019-05-15 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OZA
 
 | | Crystal structure of the phycocyanobilin-bound GAF domain from a cyanobacterial phytochrome | | 分子名称: | PHYCOCYANOBILIN, Two-component sensor histidine kinase | | 著者 | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | 登録日 | 2019-05-15 | | 公開日 | 2020-05-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.002 Å) | | 主引用文献 | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8CUH
 
 | |
8CMY
 
 | |
5HZG
 
 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | 分子名称: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | 著者 | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | 登録日 | 2016-02-02 | | 公開日 | 2016-08-03 | | 最終更新日 | 2016-08-31 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5HYW
 
 | | The crystal structure of the D3-ASK1 complex | | 分子名称: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | 著者 | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | 登録日 | 2016-02-02 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
6XIF
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 83 | | 分子名称: | GLYCEROL, Peptide 83, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Orth, P. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.774 Å) | | 主引用文献 | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XIE
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 77 | | 分子名称: | GLYCEROL, Peptide 77, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Orth, P. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XIB
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 30 | | 分子名称: | GLYCEROL, Peptide 30, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Orth, P. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.546 Å) | | 主引用文献 | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XIC
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 40 | | 分子名称: | GLYCEROL, Peptide 40, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Orth, P. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.377 Å) | | 主引用文献 | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XID
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 51 | | 分子名称: | GLYCEROL, Peptide 51, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Orth, P. | | 登録日 | 2020-06-19 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.482 Å) | | 主引用文献 | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
7QQB
 
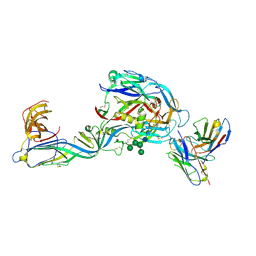 | | Crystal structure of the envelope glycoprotein complex of Puumala virus in complex with the scFv fragment of the broadly neutralizing human antibody ADI-42898 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, GLYCEROL, ... | | 著者 | Serris, A, Rey, F.A, Guardado-Calvo, P. | | 登録日 | 2022-01-07 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody.
Sci Transl Med, 15, 2023
|
|
6JCS
 
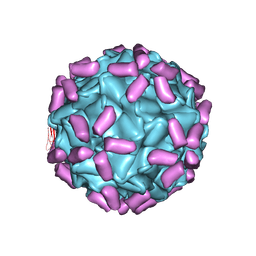 | | AAV5 in complex with AAVR | | 分子名称: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | 著者 | Lou, Z, Zhang, R. | | 登録日 | 2019-01-30 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCR
 
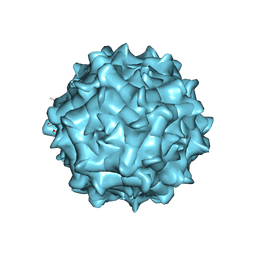 | | AAV1 in neutral condition at 3.07 Ang | | 分子名称: | Capsid protein | | 著者 | Lou, Z, Zhang, R. | | 登録日 | 2019-01-30 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCQ
 
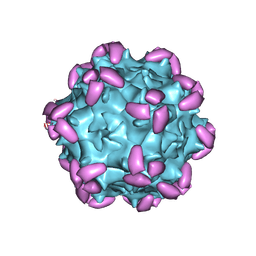 | | AAV1 in complex with AAVR | | 分子名称: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | 著者 | Lou, Z, Zhang, R. | | 登録日 | 2019-01-30 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCT
 
 | | AAV5 in neutral condition at 3.18 Ang | | 分子名称: | Capsid protein | | 著者 | Lou, Z, Zhang, R. | | 登録日 | 2019-01-30 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6K13
 
 | | Crystal Structure Basis for BmLDH Complex | | 分子名称: | L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | 著者 | Long, Y, Shen, Z. | | 登録日 | 2019-05-09 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
6K12
 
 | |
3F7O
 
 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | 分子名称: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | 著者 | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3F7M
 
 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | 分子名称: | Alkaline serine protease ver112 | | 著者 | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-09 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
6J9D
 
 | |
