4OGT
 
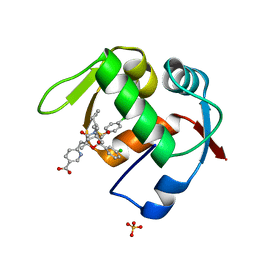 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 46 | | Descriptor: | 6-{[(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5361 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4ODF
 
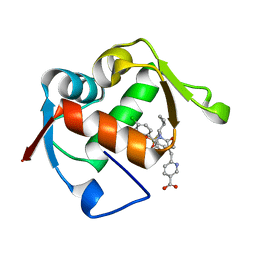 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 47 | | Descriptor: | 6-{[(2S,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2006 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
7BQA
 
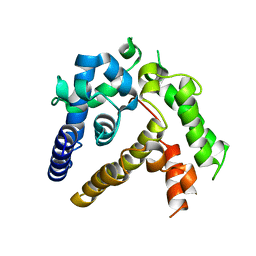 | | Crystal structure of ASFV p35 | | Descriptor: | 60 kDa polyprotein | | Authors: | Li, G.B, Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
4ODE
 
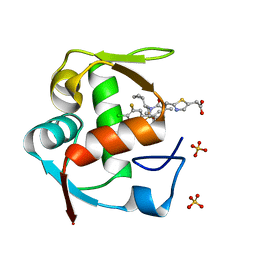 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | (2-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}-1,3-thiazol-5-yl)acetic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OGV
 
 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 49 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2S,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OGN
 
 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 3 | | Descriptor: | 6-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OBA
 
 | | Co-crystal structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Selective and Potent Morpholinone Inhibitors of the MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
4OCC
 
 | | co-crystal structure of MDM2(17-111) in complex with compound 48 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4GMB
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-402 | | Descriptor: | MM-402, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J.Med.Chem., 60, 2017
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2014-04-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
7T6T
 
 | | Structure of the human FPR1-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6V
 
 | | Structure of the human FPR2-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6U
 
 | | Structure of the human FPR2-Gi complex with CGEN-855A | | Descriptor: | B9-scFv, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6S
 
 | | Structure of the human FPR2-Gi complex with compound C43 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
6CJG
 
 | | Human dihydroorotate dehydrogenase bound to napthyridine inhibitor 46 | | Descriptor: | 2-([1,1'-biphenyl]-4-yl)-3-methyl-1,7-naphthyridine-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6CJF
 
 | | Human dihydroorotate dehydrogenase bound to 4-quinoline carboxylic acid inhibitor 43 | | Descriptor: | 2-[4-(2-chloro-6-methylpyridin-3-yl)phenyl]-6-fluoro-3-methylquinoline-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6DUM
 
 | | ALDH1A1 N121S in complex with 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (compound 13g) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | Authors: | Buchman, C.D, Hurley, T.D. | | Deposit date: | 2018-06-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy.
J.Med.Chem., 61, 2018
|
|
7XAV
 
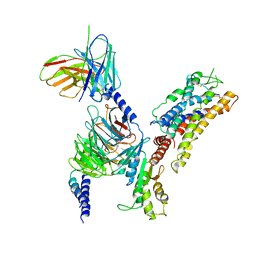 | | Structure of somatostatin receptor 2 bound with lanreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAU
 
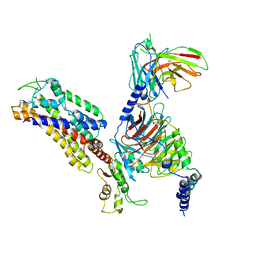 | | Structure of somatostatin receptor 2 bound with octreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
6ITC
 
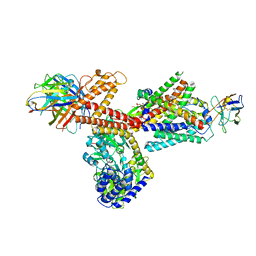 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
8HCO
 
 | | Substrate-engaged TOM complex from yeast | | Descriptor: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | Authors: | Zhou, X.Y, Yang, Y.Q, Wang, G.P, Wang, S.S. | | Deposit date: | 2022-11-02 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6KZ6
 
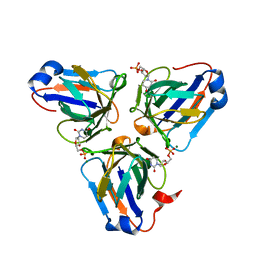 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
6MG1
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta, ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG3
 
 | | V285A Mutant of the C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
