7VFN
 
 | | Crystal structure of SdgB (SD peptide-binding form) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2G97
 
 | |
2G96
 
 | |
2G95
 
 | |
1RLD
 
 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1RLC
 
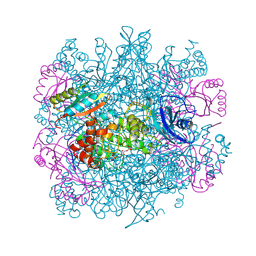 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
4KDI
 
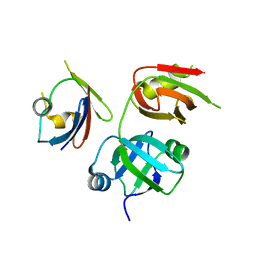 | | Crystal structure of p97/VCP N in complex with OTU1 UBXL | | Descriptor: | Transitional endoplasmic reticulum ATPase, Ubiquitin thioesterase OTU1 | | Authors: | Kim, S.J, Kim, E.E. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Ovarian Tumor Domain-containing Protein 1 (OTU1) Binding to p97/Valosin-containing Protein (VCP).
J.Biol.Chem., 289, 2014
|
|
3V6A
 
 | |
6IJR
 
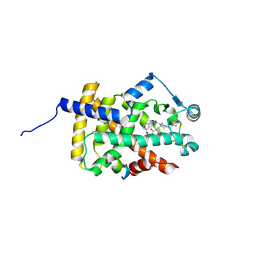 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6IJS
 
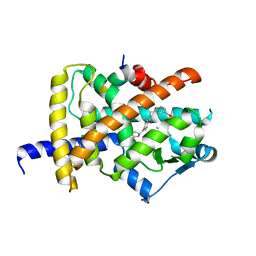 | | Human PPARgamma ligand binding domain complexed with SB1494 | | Descriptor: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6JQ7
 
 | |
1BE2
 
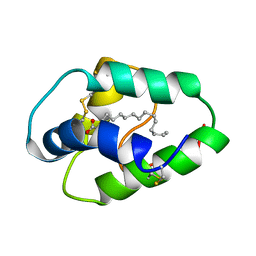 | | LIPID TRANSFER PROTEIN COMPLEXED WITH PALMITATE, NMR, 10 STRUCTURES | | Descriptor: | LIPID TRANSFER PROTEIN, PALMITIC ACID | | Authors: | Lerche, M.H, Poulsen, F.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-12-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer proteins.
Protein Sci., 7, 1998
|
|
4KDL
 
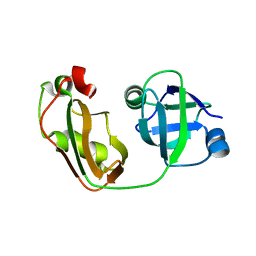 | | Crystal structure of p97/VCP N in complex with OTU1 UBXL | | Descriptor: | Transitional endoplasmic reticulum ATPase, Ubiquitin thioesterase OTU1 | | Authors: | Kim, S.J, Kim, E.E. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Ovarian Tumor Domain-containing Protein 1 (OTU1) Binding to p97/Valosin-containing Protein (VCP).
J.Biol.Chem., 289, 2014
|
|
5ZHF
 
 | | Structure of VanYB unbound | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-03-13 | | Release date: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
5ZHW
 
 | | VanYB in complex with D-Alanine-D-Alanine | | Descriptor: | COPPER (II) ION, D-ALANINE, D-alanyl-D-alanine carboxypeptidase, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-03-13 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
6A9A
 
 | | Ternary complex crystal structure of dCH with dCMP and THF | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Deoxycytidylate 5-hydroxymethyltransferase, ... | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2018-07-12 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cytosine modification mechanism revealed by the structure of a ternary complex of deoxycytidylate hydroxymethylase from bacteriophage T4 with its cofactor and substrate.
Iucrj, 6, 2019
|
|
6A9B
 
 | | T4 dCMP hydroxymethylase structure solved by I-SAD using a home source | | Descriptor: | Deoxycytidylate 5-hydroxymethyltransferase, IODIDE ION, PHOSPHATE ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2018-07-12 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A cytosine modification mechanism revealed by the structure of a ternary complex of deoxycytidylate hydroxymethylase from bacteriophage T4 with its cofactor and substrate.
Iucrj, 6, 2019
|
|
6A6A
 
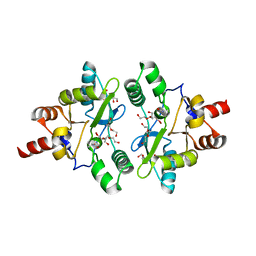 | | VanYB in complex with D-Alanine | | Descriptor: | ACETATE ION, D-ALANINE, D-alanyl-D-alanine carboxypeptidase, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
1TX6
 
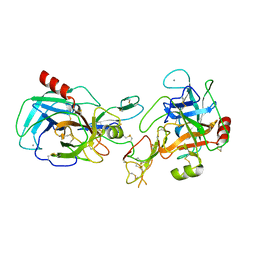 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
3SWE
 
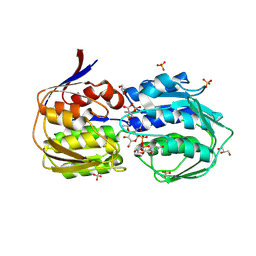 | | Haemophilus influenzae MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys117 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
2FBJ
 
 | |
2HVD
 
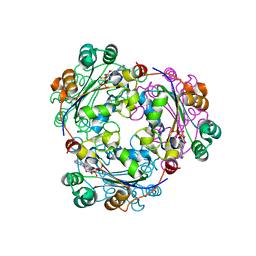 | | Human nucleoside diphosphate kinase A complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of S120G Mutant and Wild Type of Human Nucleoside Diphosphate Kinase A in Complex with ADP
J.Bioenerg.Biomembr., 38, 2006
|
|
2HVE
 
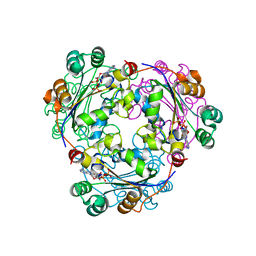 | | S120G mutant of human nucleoside diphosphate kinase A complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structures of S120G Mutant and Wild Type of Human Nucleoside Diphosphate Kinase A in Complex with ADP
J.Bioenerg.Biomembr., 38, 2006
|
|
1ZN0
 
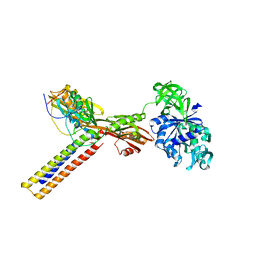 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | Descriptor: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1ZN1
 
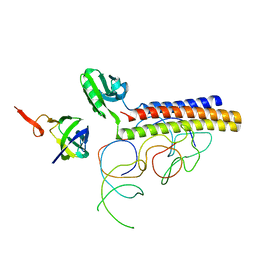 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
