5OYI
 
 | | FMDV A10 dissociated pentamer | | 分子名称: | Genome polyprotein | | 著者 | Kotecha, A, Malik, N, Stuart, D.I. | | 登録日 | 2017-09-09 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Structures of foot and mouth disease virus pentamers: Insight into capsid dissociation and unexpected pentamer reassociation.
PLoS Pathog., 13, 2017
|
|
5OWX
 
 | | Inside-out FMDV A10 capsid | | 分子名称: | Genome polyprotein | | 著者 | Kotecha, A, Malik, N, Fry, E.E, Stuart, D.I. | | 登録日 | 2017-09-04 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Structures of foot and mouth disease virus pentamers: Insight into capsid dissociation and unexpected pentamer reassociation.
PLoS Pathog., 13, 2017
|
|
5OSN
 
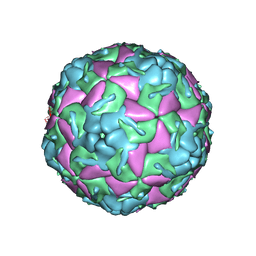 | | Crystal Structure of Bovine Enterovirus 2 determined with Serial Femtosecond X-ray Crystallography | | 分子名称: | Capsid protein, GLUTAMIC ACID, POTASSIUM ION, ... | | 著者 | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Kotecha, A, Kelly, J, Rowlands, D.J, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | 登録日 | 2017-08-17 | | 公開日 | 2017-08-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
4X3B
 
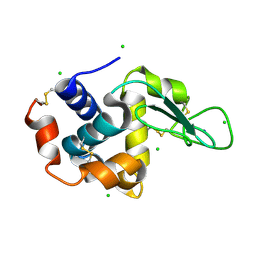 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | 登録日 | 2014-11-28 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
4X35
 
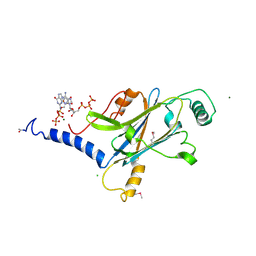 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | 登録日 | 2014-11-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
3DUZ
 
 | | Crystal structure of the postfusion form of baculovirus fusion protein GP64 | | 分子名称: | MERCURY (II) ION, Major envelope glycoprotein | | 著者 | Kadlec, J, Loureiro, S, Abrescia, N.G.A, Jones, I.M, Stuart, D.I. | | 登録日 | 2008-07-18 | | 公開日 | 2008-09-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3HHF
 
 | | Structure of CrgA regulatory domain, a LysR-type transcriptional regulator from Neisseria meningitidis. | | 分子名称: | CHLORIDE ION, Transcriptional regulator, LysR family | | 著者 | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | 登録日 | 2009-05-15 | | 公開日 | 2009-07-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
3HHG
 
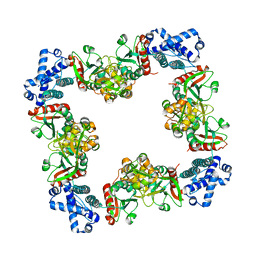 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | 分子名称: | Transcriptional regulator, LysR family | | 著者 | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | 登録日 | 2009-05-15 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
6H3B
 
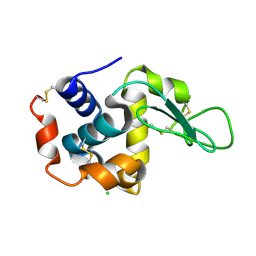 | |
6HS4
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118 | | 分子名称: | 1-[2-[3-oxidanyl-4-(4-phenyl-1~{H}-pyrazol-5-yl)phenoxy]ethyl]piperidine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Ren, J, Zhao, Y, Stuart, D.I. | | 登録日 | 2018-09-28 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
6HY0
 
 | | Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | | 分子名称: | Major Outer Capsid Protein P8, Major inner protein P1, Packaging Enzyme P4 | | 著者 | El Omari, K, Ilca, S.L, Stuart, D.I, Huiskonen, J.T. | | 登録日 | 2018-10-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus.
Nature, 570, 2019
|
|
6HRO
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118a | | 分子名称: | 1-[2-[4-[4-(4-chlorophenyl)-3-methyl-1~{H}-pyrazol-5-yl]-3-oxidanyl-phenoxy]ethyl]piperidin-1-ium-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Ren, J, Zhao, Y, Stuart, D.I. | | 登録日 | 2018-09-27 | | 公開日 | 2019-02-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
6I2K
 
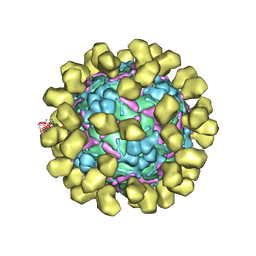 | | Structure of EV71 complexed with its receptor SCARB2 | | 分子名称: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | 登録日 | 2018-11-01 | | 公開日 | 2018-11-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
1RT3
 
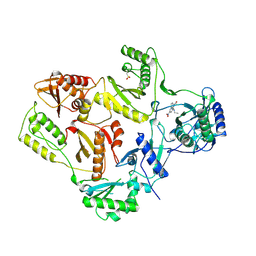 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | 分子名称: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Stammers, D.K, Stuart, D.I. | | 登録日 | 1998-06-29 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6J7V
 
 | |
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | 分子名称: | FAB Heavy chain, FAB Light chain | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.907 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1ALC
 
 | |
4Q4F
 
 | | Crystal structure of LIMP-2 (space group C2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | 登録日 | 2014-04-14 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
4Q4B
 
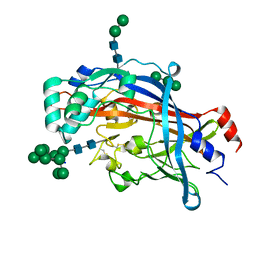 | | Crystal structure of LIMP-2 (space group C2221) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | 著者 | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | 登録日 | 2014-04-14 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
4QPG
 
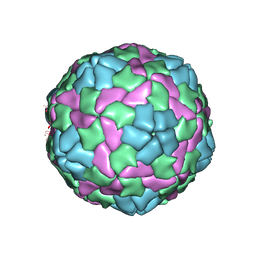 | | Crystal structure of empty hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QPI
 
 | | Crystal structure of hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4S1K
 
 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 100 K | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | 著者 | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | 登録日 | 2015-01-14 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
4S1L
 
 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 298 K | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, polyhedrin | | 著者 | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | 登録日 | 2015-01-14 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
4CDU
 
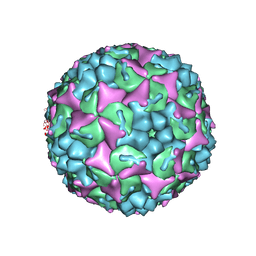 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-06 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
