8A97
 
 | | ROOM TEMPERATURE CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) UNDER XENON PRESSURE (30 bar) | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, D(-)-TARTARIC ACID, XENON | | Authors: | Bui, S, Prange, T, Steiner, R.A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
5FJY
 
 | | Crystal structure of mouse kinesin light chain 2 (residues 161-480) | | Descriptor: | KINESIN LIGHT CHAIN 2, UNKNOWN PEPTIDE | | Authors: | Pernigo, S, Yip, Y.Y, Sanger, A, Xu, M, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Light Chains of Kinesin-1 are Autoinhibited.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7OKZ
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL- QUINOLIN-4(1H)-ONE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
7OJM
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2021-05-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
6FIZ
 
 | | Crystal Structure of CNG mimicking NaK-EAPP mutant (T67A) cocrystallized with K+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, POTASSIUM ION, ... | | Authors: | Napolitano, L.M.R, De March, M, Steiner, R.A, Onesti, S. | | Deposit date: | 2018-01-19 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structure of CNG mimicking NaK-EAPP mutant (T67A) cocrystallized with K+
To Be Published
|
|
2Y9R
 
 | |
4UOW
 
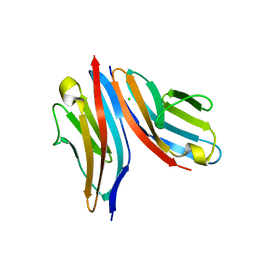 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
1XCS
 
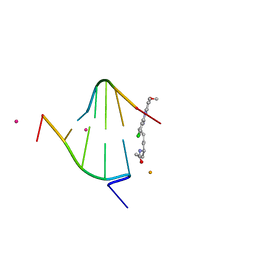 | | structure of oligonucleotide/drug complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-[(5-(ACETYLAMINO)-6-{[(1S,4R)-8-AMINO-4-[((2R)-6-AMINO-2-{2-[(1S)-5-AMINO-1-FORMYLPENTYL]HYDRAZINO}HEXANOYL)AMINO]-1-(4-AMINOBUTYL)-2,3-DIOXOOCTYL]AMINO}-6-OXOHEXYL)AMINO]-6-CHLORO-2-METHOXYACRIDINIUM, BARIUM ION, ... | | Authors: | Valls, N, Steiner, R.A, Wright, G, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Variable role of ions in two drug intercalation complexes of DNA
J.Biol.Inorg.Chem., 10, 2005
|
|
1XCU
 
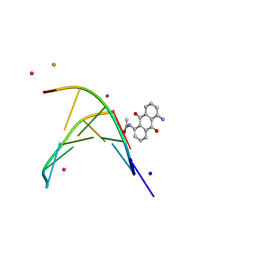 | | oligonucleotid/drug complex | | Descriptor: | 1,5-BIS[3-(DIETHYLAMINO)PROPIONAMIDO]ANTHRACENE-9,10-DIONE, 5'-D(*CP*GP*TP*AP*CP*G)-3', BARIUM ION, ... | | Authors: | Valls, N, Steiner, R.A, Wright, G, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variable role of ions in two drug intercalation complexes of DNA
J.Biol.Inorg.Chem., 10, 2005
|
|
4CFS
 
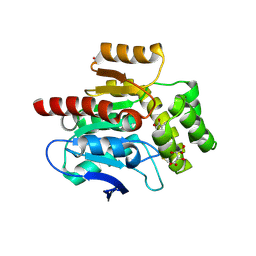 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE | | Descriptor: | 1-H-3-HYDROXY-4-OXOQUINALDINE 2,4-DIOXYGENASE, 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Origin of the Proton-Transfer Step in the Cofactor-Free 1-H-3-Hydroxy-4-Oxoquinaldine 2,4- Dioxygenase: Effect of the Basicity of an Active Site His Residue.
J.Biol.Chem., 289, 2014
|
|
7A0L
 
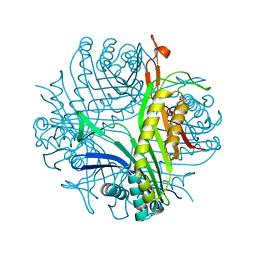 | | Joint neutron/X-ray room temperature structure of perdeuterated Aspergillus flavus urate oxidase in complex with the 8-azaxanthine inhibitor and catalytic water bound in the peroxo hole | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | McGregor, L, Bui, S, Blakeley, M.P, Steiner, R.A. | | Deposit date: | 2020-08-09 | | Release date: | 2020-12-09 | | Last modified: | 2021-02-10 | | Method: | NEUTRON DIFFRACTION (1.33 Å), X-RAY DIFFRACTION | | Cite: | Joint neutron/X-ray crystal structure of a mechanistically relevant complex of perdeuterated urate oxidase and simulations provide insight into the hydration step of catalysis.
Iucrj, 8, 2021
|
|
5FM4
 
 | |
5FM8
 
 | |
5FM5
 
 | |
2WP3
 
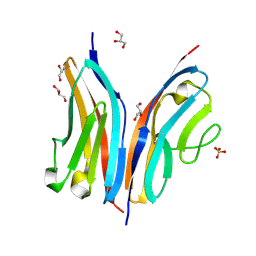 | | Crystal structure of the Titin M10-Obscurin like 1 Ig complex | | Descriptor: | GLYCEROL, OBSCURIN-LIKE PROTEIN 1, SULFATE ION, ... | | Authors: | Pernigo, S, Fuzukawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-08-02 | | Release date: | 2010-02-16 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WWK
 
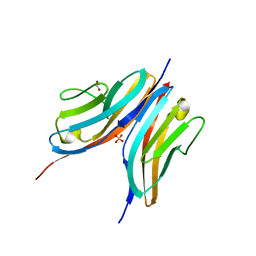 | | Crystal structure of the Titin M10-Obscurin like 1 Ig F17R mutant complex | | Descriptor: | GLYCEROL, OBSCURIN-LIKE PROTEIN 1, SULFATE ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-10-25 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WWM
 
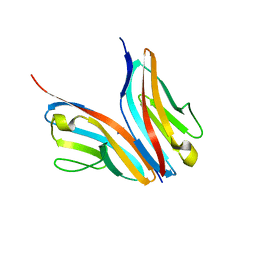 | | Crystal structure of the Titin M10-Obscurin like 1 Ig complex in space group P1 | | Descriptor: | OBSCURIN-LIKE PROTEIN 1, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1S23
 
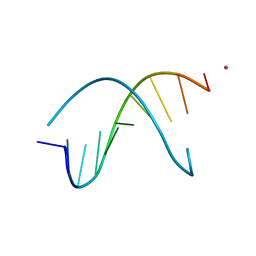 | | Crystal Structure Analysis of the B-DNA Decamer CGCAATTGCG | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*TP*TP*GP*CP*G)-3', COBALT (II) ION | | Authors: | Valls, N, Wright, G, Steiner, R.A, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA variability in five crystal structures of d(CGCAATTGCG).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1JUH
 
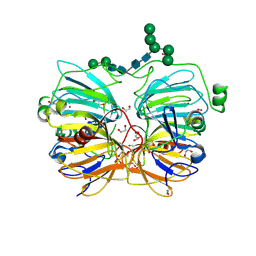 | | Crystal Structure of Quercetin 2,3-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Schroeter, K.H, Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-08-24 | | Release date: | 2002-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus.
Structure, 10, 2002
|
|
3ZFW
 
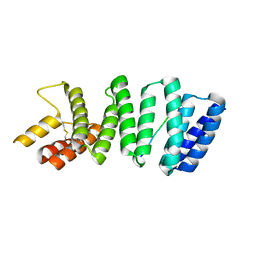 | | Crystal structure of the TPR domain of kinesin light chain 2 in complex with a tryptophan-acidic cargo peptide | | Descriptor: | KINESIN LIGHT CHAIN 2, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY M MEMBER 2 | | Authors: | Pernigo, S, Lamprecht, A, Steiner, R.A, Dodding, M.P. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Kinesin-1:Cargo Recognition.
Science, 340, 2013
|
|
4C4K
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | 1,2-ETHANEDIOL, OBSCURIN, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
4CW2
 
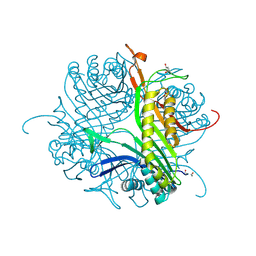 | | Crystal structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-metyl uric acid (X-ray dose, 2.5 kGy) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, URICASE | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-29 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Direct evidence for a peroxide intermediate and a reactive enzyme-substrate-dioxygen configuration in a cofactor-free oxidase.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4D17
 
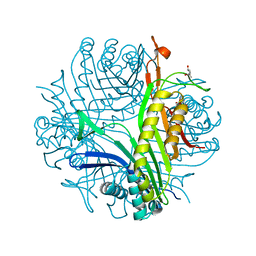 | | Crystal structure of cofactor-free urate oxidase in complex with its 5-peroxoisourate intermediate (X-ray dose, 106 kGy) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(HYDRO)PEROXOISOURATE, OXYGEN MOLECULE, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-05 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Direct evidence for a peroxide intermediate and a reactive enzyme-substrate-dioxygen configuration in a cofactor-free oxidase.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4CW3
 
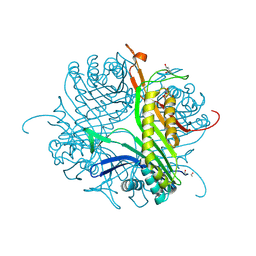 | | Crystal structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-metyl uric acid (X-ray dose, 665 kGy) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, 9-METHYL URIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-29 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Direct evidence for a peroxide intermediate and a reactive enzyme-substrate-dioxygen configuration in a cofactor-free oxidase.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4D12
 
 | |
