7PGT
 
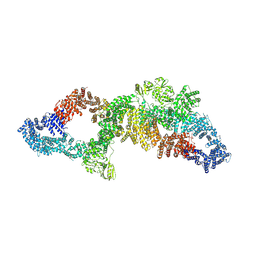 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGS
 
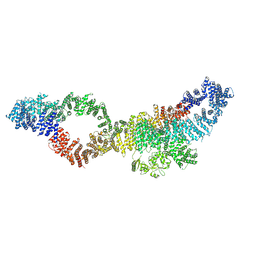 | | Consensus structure of human Neurofibromin isoform 2 | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGR
 
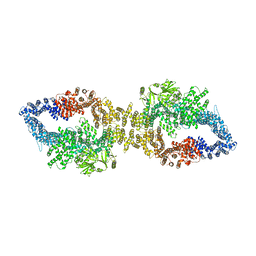 | | The structure of human neurofibromin isoform 2 in closed conformation | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGU
 
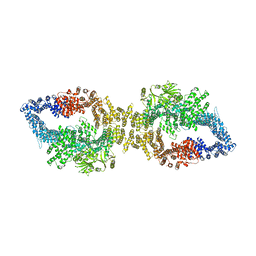 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
1YK3
 
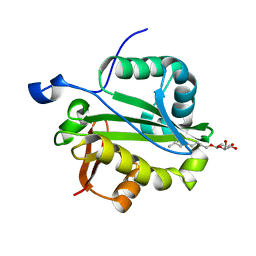 | | Crystal structure of Rv1347c from Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv1347c/MT1389, octyl beta-D-glucopyranoside | | Authors: | Card, G.L, Peterson, N.A, Smith, C.A, Rupp, B, Schick, B.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-01-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis
J.Biol.Chem., 280, 2005
|
|
1YNK
 
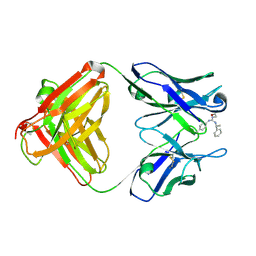 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeteners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-[((R)-{[4-(AMINOMETHYL)PHENYL]AMINO}{[(1R)-1-PHENYLETHYL]AMINO}METHYL)AMINO]ETHANE-1,1-DIOL, Ig gamma heavy chain, immunoglobulin kappa light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
1YNL
 
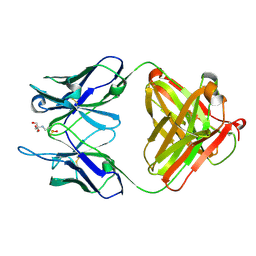 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeterners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Ig gamma heavy chain, Ig gamma light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
1UPI
 
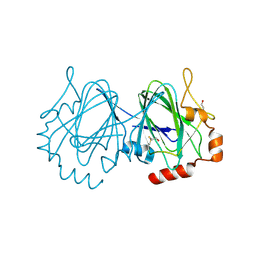 | | Mycobacterium tuberculosis rmlC epimerase (Rv3465) | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Kim, C.-Y, Naranjo, C, Waldo, G.S, Lekin, T, Segelke, B.W, Kantardjieff, K.A, Zemla, A, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-10-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium Tuberculosis Rmlc Epimerase (Rv3465): A Promising Drug-Target Structure in the Rhamnose Pathway
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1W30
 
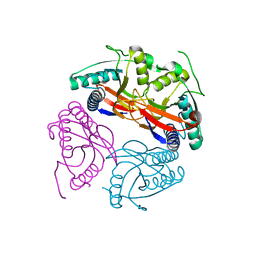 | | PyrR of Mycobacterium Tuberculosis as a potential drug target | | Descriptor: | PYRR BIFUNCTIONAL PROTEIN | | Authors: | Kantardjieff, K.A, Vasquez, C, Castro, P, Warfel, N.N, Rho, B.-S, Lekin, T, Kim, C.-Y, Segelke, B.W, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pyrr (Rv1379) from Mycobacterium Tuberculosis: A Persistence Gene and Protein Drug Target
Acta Crystallogr.,Sect.D, 61, 2005
|
|
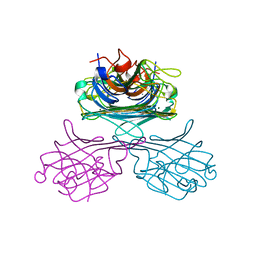
1E1H
 
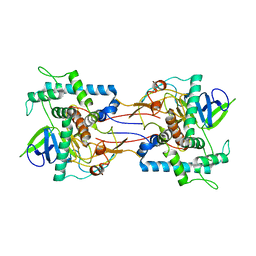 | |
1GKB
 
 | | CONCANAVALIN A, NEW CRYSTAL FORM | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MAGNESIUM ION, ... | | Authors: | Kantardjieff, K, Rupp, B, Hoechtl, P, Segelke, B. | | Deposit date: | 2001-08-10 | | Release date: | 2001-08-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concanavalin a in a Dimeric Crystal Form: Revisiting Structural Accuracy and Molecular Flexibility
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GS9
 
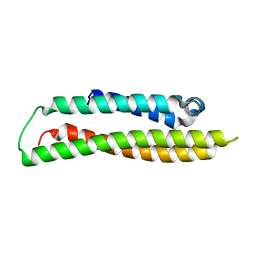 | | Apolipoprotein E4, 22k domain | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Verderame, J.R, Kantardjieff, K, Segelke, B, Weisgraber, K, Rupp, B. | | Deposit date: | 2002-01-02 | | Release date: | 2003-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the 22K Domain of Human Apolipoprotein E4
To be Published
|
|
1BPI
 
 | |
1DYQ
 
 | | STAPHYLOCOCCAL ENTEROTOXIN A MUTANT VACCINE | | Descriptor: | Enterotoxin type A, SULFATE ION, ZINC ION | | Authors: | Krupka, H.I, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-02-05 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Abrogated Binding between Staphylococcal Enterotoxin a Superantigen Vaccine and Mhc-II?
Protein Sci., 11, 2002
|
|
8PB6
 
 | | PsiM in complex with SAH and baeocystin | | Descriptor: | Baeocystin, CHLORIDE ION, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8PB4
 
 | | PsiM in complex with SAH and norbaeocystin, orthorhombic crystal form | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8PB8
 
 | | PsiM in complex with SAH | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8PB7
 
 | | PsiM in complex with sinefungin and baeocystin | | Descriptor: | Baeocystin, CHLORIDE ION, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8PB3
 
 | | PsiM in complex with SAH and norbaeocystin, monoclinic crystal form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Norbaeocystin, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8PB5
 
 | | PsiM in complex with sinefungin and norbaeocystin | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
8QXQ
 
 | | PsiM in complex with SAH and psilocybin | | Descriptor: | CHLORIDE ION, Psilocybin synthase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-10-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VF2
 
 | | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VGA
 
 | | Alternative model for Fab 36-65 | | Descriptor: | Fab 36-65 heavy chain, Fab 36-65 light chain, TRIETHYLENE GLYCOL | | Authors: | Stanfield, R.L, Rupp, B, Wlodawer, A, Dauter, Z, Porebski, P.J, Minor, W, Jaskolski, M, Pozharski, E, Weichenberger, C.X. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-06 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
