6JC0
 
 | |
6L42
 
 | |
6BUI
 
 | |
6BUG
 
 | |
5YJZ
 
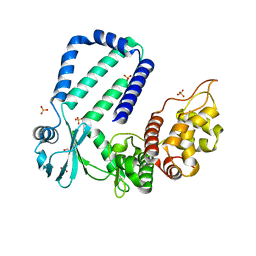 | | The native crystal structure of Rv3197 from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Probable conserved ATP-binding protein ABC transporter, SULFATE ION | | Authors: | Zhang, Q.Q, Rao, Z.H. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK1
 
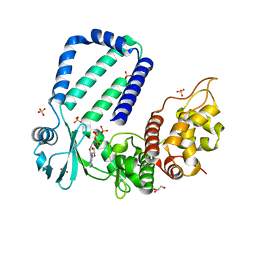 | | The complex structure of Rv3197-AMPPNP from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK2
 
 | |
5YK0
 
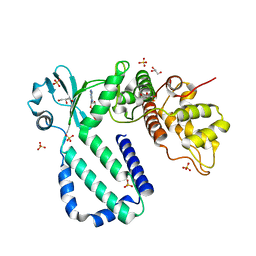 | | The complex structure of Rv3197-ADP from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
7BQA
 
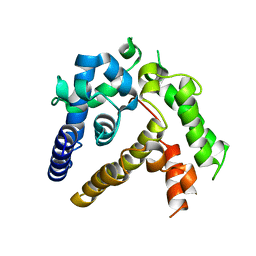 | | Crystal structure of ASFV p35 | | Descriptor: | 60 kDa polyprotein | | Authors: | Li, G.B, Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
7CAG
 
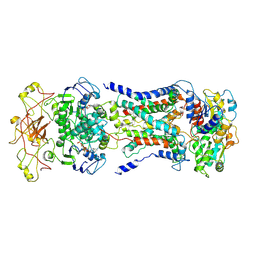 | | Mycobacterium smegmatis LpqY-SugABC complex in the catalytic intermediate state | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
6AKS
 
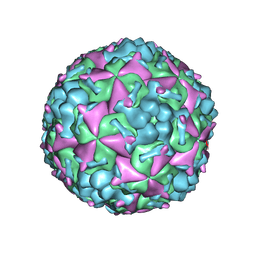 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
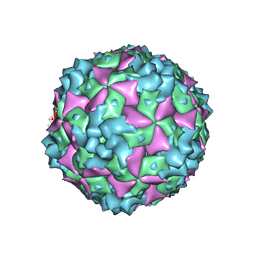 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
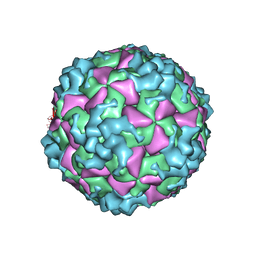 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6JTG
 
 | |
8GPY
 
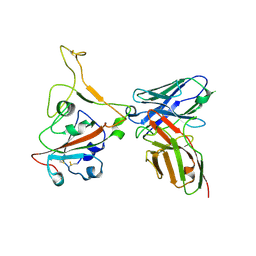 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
3IWM
 
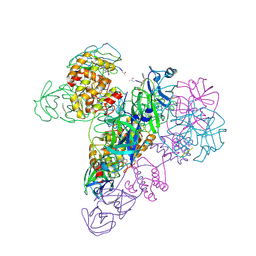 | | The octameric SARS-CoV main protease | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | Deposit date: | 2009-09-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
6KZ6
 
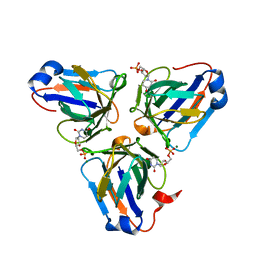 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
5YEW
 
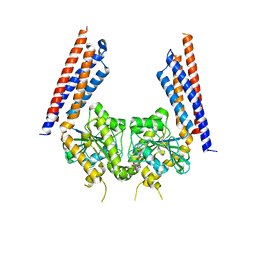 | | Structural basis for GTP hydrolysis and conformational change of mitofusin 1 in mediating mitochondrial fusion | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, L, Qi, Y, Huang, X, Yu, C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion
Nat. Struct. Mol. Biol., 25, 2018
|
|
4JR6
 
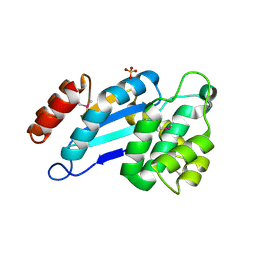 | | Crystal structure of DsbA from Mycobacterium tuberculosis (reduced) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
4JOD
 
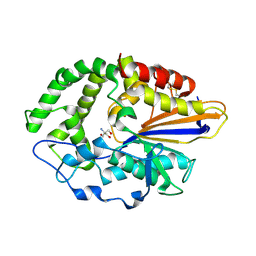 | |
4JOB
 
 | |
4JR4
 
 | | Crystal structure of Mtb DsbA (Oxidized) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
4JOC
 
 | |
4MG3
 
 | | Crystal Structural Analysis of 2A Protease from Coxsackievirus A16 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protease 2A, ... | | Authors: | Sun, Y, Wang, X, Dang, M, Yuan, S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | An open conformation determined by a structural switch for 2A protease from coxsackievirus A16.
Protein Cell, 4, 2013
|
|
