6IGL
 
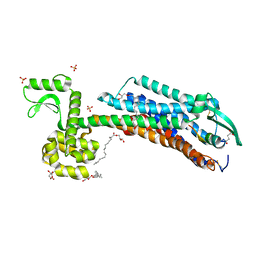 | | Crystal Structure of human ETB receptor in complex with IRL1620 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | 著者 | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | 登録日 | 2018-09-25 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
6IRW
 
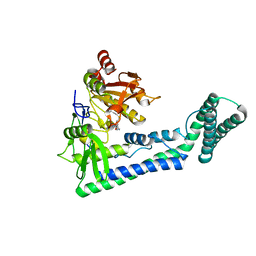 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IDP
 
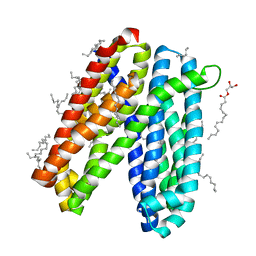 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the straight form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | 著者 | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | 登録日 | 2018-09-11 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.205 Å) | | 主引用文献 | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IU3
 
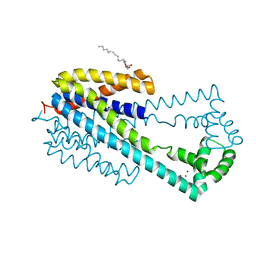 | | Crystal structure of iron transporter VIT1 with zinc ions | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | 著者 | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-27 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IDR
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | 著者 | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | 登録日 | 2018-09-11 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IRX
 
 | |
6IU6
 
 | | Crystal structure of cytoplasmic metal binding domain with nickel ions | | 分子名称: | NICKEL (II) ION, VIT1, ZINC ION | | 著者 | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-27 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IRY
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | 分子名称: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | 著者 | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | 登録日 | 2016-01-24 | | 公開日 | 2016-04-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
6JOO
 
 | | Crystal structure of Corynebacterium diphtheriae Cas9 in complex with sgRNA and target DNA | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated protein,CRISPR-associated endonuclease Cas9, Guide RNA, ... | | 著者 | Hirano, S, Ishitani, R, Nishimasu, H, Nureki, O. | | 登録日 | 2019-03-22 | | 公開日 | 2019-04-17 | | 最終更新日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9.
Nat Commun, 10, 2019
|
|
6JMQ
 
 | | LAT1-CD98hc complex bound to MEM-108 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | 著者 | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | 登録日 | 2019-03-13 | | 公開日 | 2019-06-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JKD
 
 | | Crystal structure of tetrameric PepTSo2 in I4 space group | | 分子名称: | Proton:oligopeptide symporter POT family | | 著者 | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | 登録日 | 2019-02-28 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JKC
 
 | | Crystal structure of tetrameric PepTSo2 in P4212 space group | | 分子名称: | Proton:oligopeptide symporter POT family | | 著者 | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | 登録日 | 2019-02-28 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JMR
 
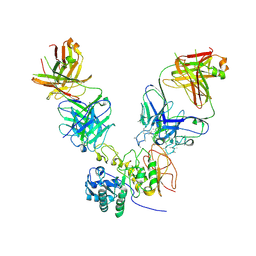 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | 著者 | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | 登録日 | 2019-03-13 | | 公開日 | 2019-06-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5I20
 
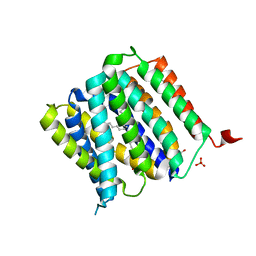 | | Crystal structure of protein | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | 著者 | Ishitani, R, Nureki, O. | | 登録日 | 2016-02-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
6K7H
 
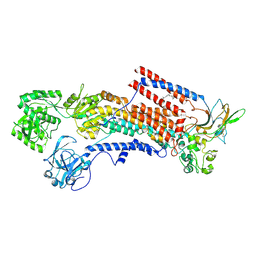 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-06-07 | | 公開日 | 2019-08-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7N
 
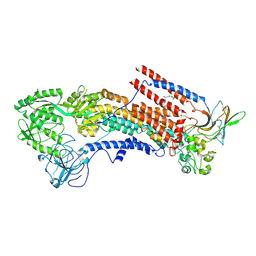 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-06-07 | | 公開日 | 2019-08-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7I
 
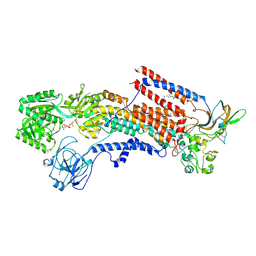 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-06-07 | | 公開日 | 2019-08-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7J
 
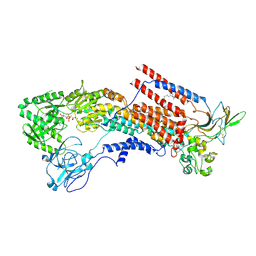 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-06-07 | | 公開日 | 2019-08-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K4J
 
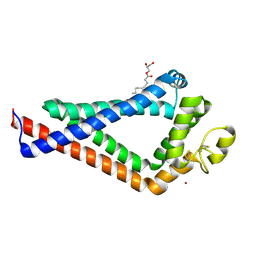 | | Crystal Structure of the the CD9 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | 著者 | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | 登録日 | 2019-05-24 | | 公開日 | 2020-05-13 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
4YSC
 
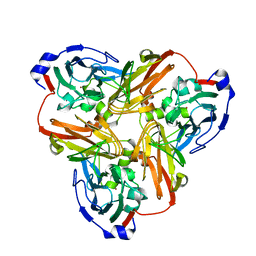 | | Completely oxidized structure of copper nitrite reductase from Alcaligenes faecalis | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-03-17 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YSR
 
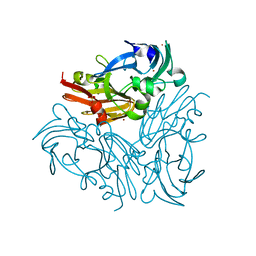 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.6 MGy | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | 著者 | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-03-17 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSE
 
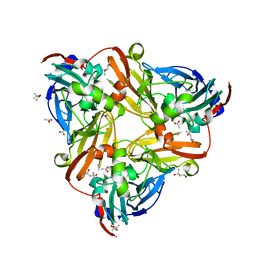 | | High resolution synchrotron structure of copper nitrite reductase from Alcaligenes faecalis | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, COPPER (II) ION, ... | | 著者 | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-03-17 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YZI
 
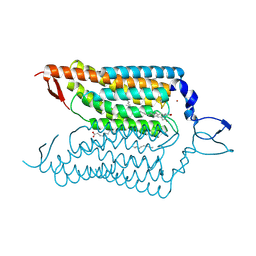 | | Crystal structure of blue-shifted channelrhodopsin mutant (T198G/G202A) | | 分子名称: | OLEIC ACID, RETINAL, Sensory opsin A,Archaeal-type opsin 2, ... | | 著者 | Kato, H.E, Kamiya, M, Ishitani, R, Hayashi, S, Nureki, O. | | 登録日 | 2015-03-25 | | 公開日 | 2015-05-27 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Atomistic design of microbial opsin-based blue-shifted optogenetics tools.
Nat Commun, 6, 2015
|
|
7V6B
 
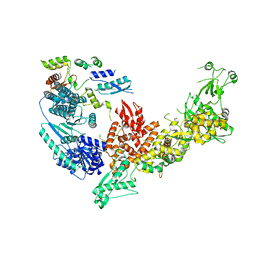 | | Structure of the Dicer-2-R2D2 heterodimer | | 分子名称: | Dicer-2, isoform A, R2D2 | | 著者 | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | 登録日 | 2021-08-20 | | 公開日 | 2022-03-23 | | 最終更新日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
