1KA5
 
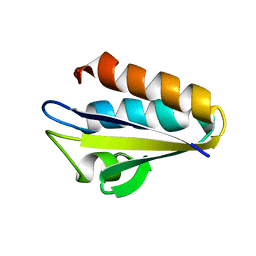 | | Refined Solution Structure of Histidine Containing Phosphocarrier Protein from Staphyloccocus aureus | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Maurer, T, Meier, S, Hengstenberg, W, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2001-10-31 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus aureus and characterization of its interaction with the bifunctional HPr kinase/phosphorylase
J.Bacteriol., 186, 2004
|
|
1QFR
 
 | | NMR SOLUTION STRUCTURE OF PHOSPHOCARRIER PROTEIN HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Maurer, T, Doeker, R, Goerler, A, Hengstenberg, W, Kalbitzer, H.R. | | Deposit date: | 1999-04-13 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution.
Eur.J.Biochem., 268, 2001
|
|
4DSO
 
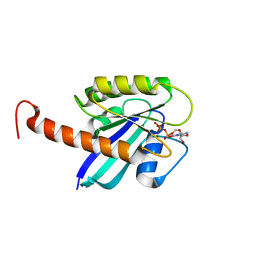 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
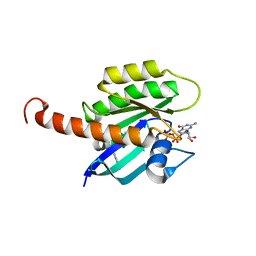 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DST
 
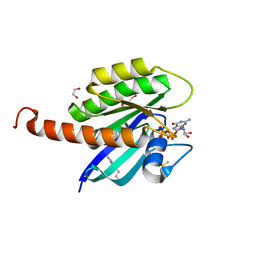 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2B3A
 
 | | Solution structure of the Ras-binding domain of the Ral Guanosine Dissociation Stimulator | | Descriptor: | Ral guanine nucleotide dissociation stimulator | | Authors: | Gronwald, W, Maurer, T, Fuechsl, R, Wohlgemuth, S, Herrmann, C, Kalbitzer, H.R. | | Deposit date: | 2005-09-20 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | New insights into binding of the possible cancer target RalGDS
To be Published
|
|
2K13
 
 | | Solution NMR Structure of the Leech Protein Saratin, a Novel Inhibitor of Haemostasis | | Descriptor: | Saratin | | Authors: | Gronwald, W, Bomke, J, Maurer, T, Wisotzki, B, Huber, F, Schumann, F, Kremer, W, Frech, M, Kalbitzer, H.R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the leech protein saratin and characterization of its binding to collagen
J.Mol.Biol., 381, 2008
|
|
1A7M
 
 | | LEUKAEMIA INHIBITORY FACTOR CHIMERA (MH35-LIF), NMR, 20 STRUCTURES | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Hinds, M.G, Maurer, T, Zhang, J.-G, Nicola, N.A, Norton, R.S. | | Deposit date: | 1998-03-16 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leukemia inhibitory factor.
J.Biol.Chem., 273, 1998
|
|
1P6Q
 
 | | NMR Structure of the Response regulator CheY2 from Sinorhizobium meliloti, complexed with Mg++ | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the inactive and BeF3-activated response regulator CheY2.
J.Mol.Biol., 338, 2004
|
|
1P6U
 
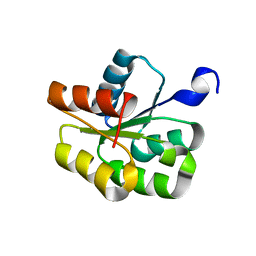 | | NMR structure of the BeF3-activated structure of the response regulator Chey2-Mg2+ from Sinorhizobium meliloti | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Inactive and BeF(3)-activated Response Regulator CheY2
J.Biol.Chem., 338, 2004
|
|
1UB1
 
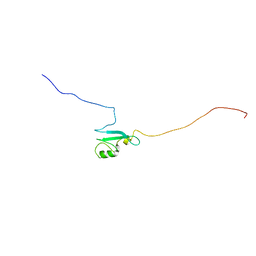 | | Solution structure of the matrix attachment region-binding domain of chicken MeCP2 | | Descriptor: | attachment region binding protein | | Authors: | Heitmann, B, Maurer, T, Weitzel, J.M, Stratling, W.H, Kalbitzer, H.R, Brunner, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the matrix attachment region-binding domain of chicken MeCP2
EUR.J.BIOCHEM., 270, 2003
|
|
5JTJ
 
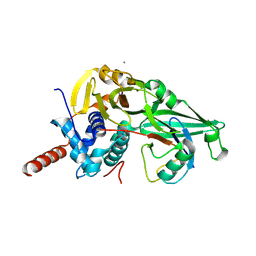 | | USP7CD-CTP in complex with Ubiquitin | | Descriptor: | CALCIUM ION, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7,Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
5JTV
 
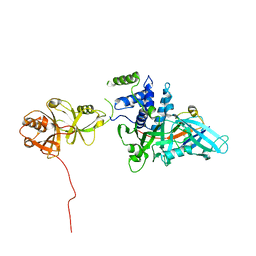 | | USP7CD-UBL45 in complex with Ubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2016-05-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Molecular Understanding of USP7 Substrate Recognition and C-Terminal Activation.
Structure, 24, 2016
|
|
5UQV
 
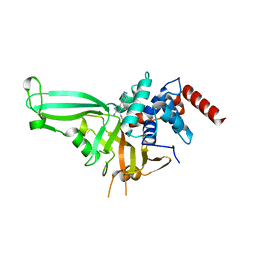 | |
5UQX
 
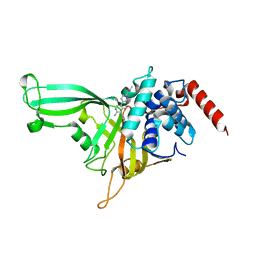 | |
5J7T
 
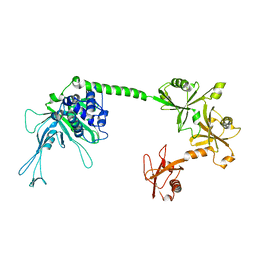 | |
3TMO
 
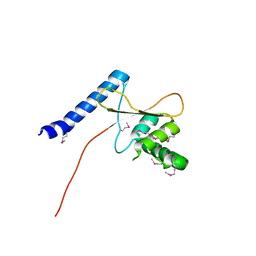 | | The catalytic domain of human deubiquitinase DUBA | | Descriptor: | OTU domain-containing protein 5 | | Authors: | Yin, J, Bosanac, I, Ma, X, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TMP
 
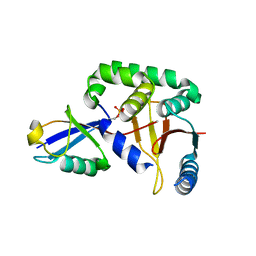 | | The catalytic domain of human deubiquitinase DUBA in complex with ubiquitin aldehyde | | Descriptor: | OTU domain-containing protein 5, Polyubiquitin-C | | Authors: | Ma, X, Yin, J, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5WHC
 
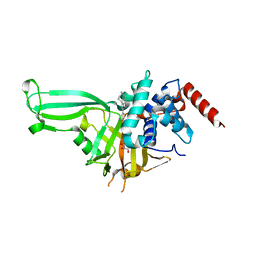 | | USP7 in complex with Cpd2 (4-(3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl)phenol) | | Descriptor: | 4-[3-(1-methylpiperidin-4-yl)-1,2,4-oxadiazol-5-yl]phenol, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-07-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of Ubiquitin Specific Protease 7 (USP7) Using Integrated NMR and in Silico Techniques.
J. Med. Chem., 60, 2017
|
|
5VZY
 
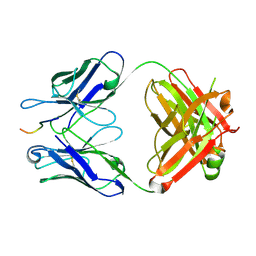 | | Crystal structure of crenezumab Fab in complex with Abeta | | Descriptor: | Amyloid beta A4 protein, Crenezumab Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Crenezumab Fab light chain,Immunoblobulin light chain | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
5VZX
 
 | | Crystal structure of crenezumab Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Crenezumab Fab heavy chain, ... | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
6BPP
 
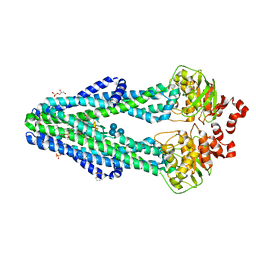 | | E. coli MsbA in complex with LPS and inhibitor G092 | | Descriptor: | (2E)-3-{6-[(1S)-1-(3-amino-2,6-dichlorophenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
6BPL
 
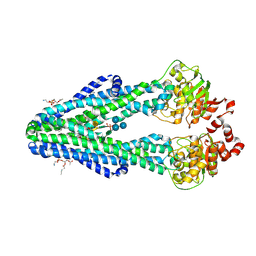 | | E. coli MsbA in complex with LPS and inhibitor G907 | | Descriptor: | (2E)-3-{6-[(1S)-1-(2-chloro-6-cyclopropylphenyl)ethoxy]-4-cyclopropylquinolin-3-yl}prop-2-enoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-amino-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Ho, H, Koth, C.M, Payandeh, J. | | Deposit date: | 2017-11-23 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural basis for dual-mode inhibition of the ABC transporter MsbA.
Nature, 557, 2018
|
|
2QMV
 
 | |
