3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMS
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
7U3P
 
 | | [3T7] Self-assembling tensegrity triangle with three turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.06 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Q
 
 | | [4T7] Self-assembling tensegrity triangle with four turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (29-MER), DNA (42-MER), DNA (5'-D(P*AP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P, Huang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.32 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
4C6R
 
 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPS4 | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
5ZCJ
 
 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
7XKS
 
 | | Crystal structure of an alkaline pectate lyase from Bacillus clausii | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase | | Authors: | Zhou, C, Zheng, Y.Y, Liu, W.D, Ma, Y. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an Alkaline Pectate Lyase and Rational Engineering with Improved Thermo-Alkaline Stability for Efficient Ramie Degumming.
Int J Mol Sci, 24, 2022
|
|
5ZQY
 
 | | Crystal structure of a poly(ADP-ribose) glycohydrolase | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, M, Yuan, Z, Ma, Y, Wang, J, Liu, X. | | Deposit date: | 2018-04-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation.
J. Biol. Chem., 293, 2018
|
|
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
7RVR
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI18 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVV
 
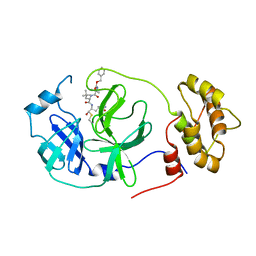 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW1
 
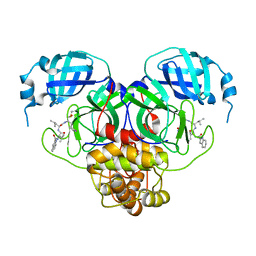 | |
7RVY
 
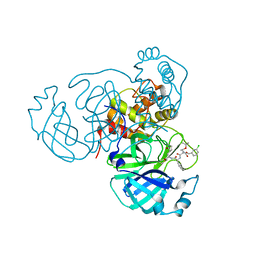 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI25 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVS
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVX
 
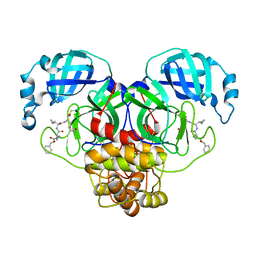 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI24 | | Descriptor: | 3C-like proteinase, benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVM
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI11 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVN
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
