3VC8
 
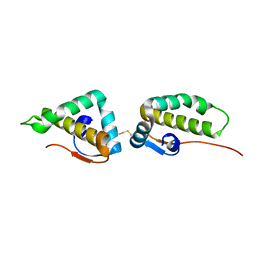 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3WDJ
 
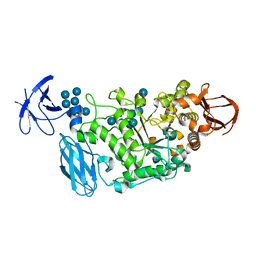 | | Crystal structure of Pullulanase complexed with maltotetraose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDH
 
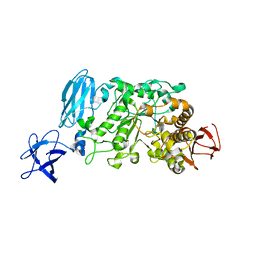 | | Crystal structure of Pullulanase from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDI
 
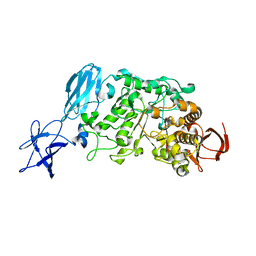 | | Crystal structure of Pullulanase complexed with maltotriose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3AXD
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6LNA
 
 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
6M1I
 
 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
3F3P
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21212 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F3F
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P21 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F3G
 
 | | Crystal structure of the nucleoporin pair Nup85-Seh1, space group P212121 | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Debler, E.W, Hseo, H, Ma, Y, Blobel, G, Hoelz, A. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A fence-like coat for the nuclear pore membrane.
Mol.Cell, 32, 2008
|
|
3F7F
 
 | | Structure of Nup120 | | Descriptor: | MERCURY (II) ION, Nucleoporin NUP120 | | Authors: | Seo, H.S, Ma, Y, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-11-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8BDG
 
 | | Tubulin-taxane-2b complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDE
 
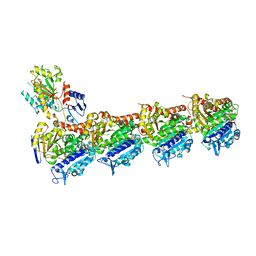 | | Tubulin-baccatin III complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDF
 
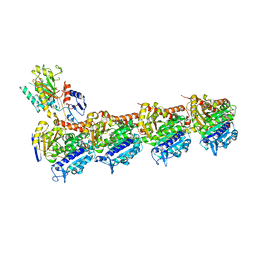 | | Tubulin-taxane-2a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
3H7N
 
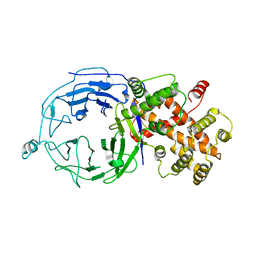 | | Structure of Nup120 | | Descriptor: | Nucleoporin NUP120 | | Authors: | Seo, H.S, Ma, Y, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2009-04-27 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6LH4
 
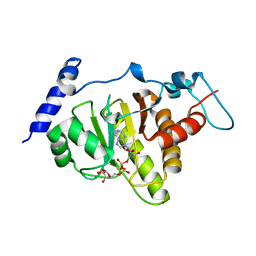 | | Crystal structural of MacroD1-ADPr complex | | Descriptor: | ADP-ribose glycohydrolase MACROD1, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yang, X, Ma, Y, Li, Y. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular basis for the MacroD1-mediated hydrolysis of ADP-ribosylation.
DNA Repair (Amst), 94, 2020
|
|
5U6A
 
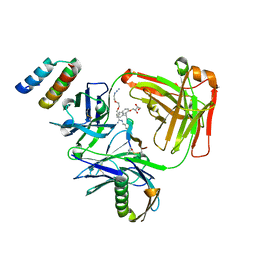 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-PEG3-MEDITOPE | | Descriptor: | Heavy Chain, Immunoglobulin G binding protein A, Light Chain, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-PEG3-Meditope
To Be Published
|
|
5U5M
 
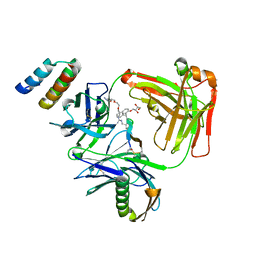 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-MEDITOPE | | Descriptor: | AZIDO-PEG4-MEDITOPE, Immunoglobulin G binding protein A, MEMAB TRASTUZUMAB, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-Meditope
To Be Published
|
|
4DIN
 
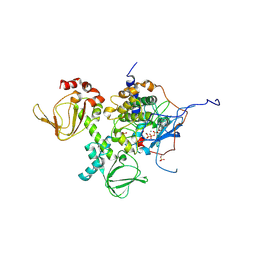 | | Novel Localization and Quaternary Structure of the PKA RI beta Holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Ilouz, R, Bubis, J, Wu, J, Yim, Y.Y, Deal, M.S, Kornev, A.P, Ma, Y, Blumenthal, D.K, Taylor, S.S. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Localization and quaternary structure of the PKA RI Beta holoenzyme
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
3AMD
 
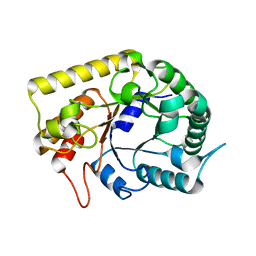 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMR
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CALCIUM ION, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
