4ALI
 
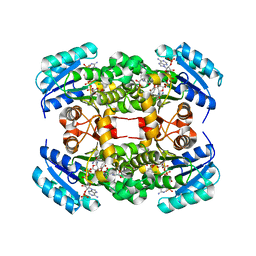 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
5I83
 
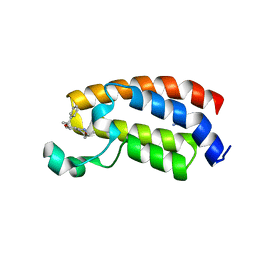 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02773986 | | Descriptor: | (4R)-4-methyl-7-[(1R)-1-phenylethoxy]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein, THIOCYANATE ION | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I86
 
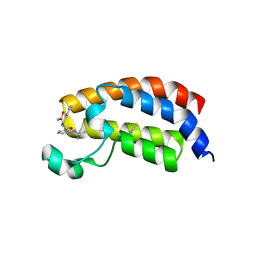 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I89
 
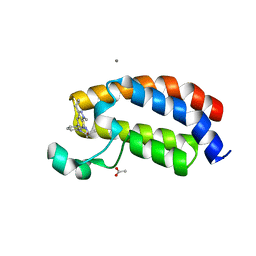 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02857790 | | Descriptor: | (4R)-6-(3-cyclopropyl-1-methyl-1H-indazol-5-yl)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Setser, J.W, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I8G
 
 | | CBP in complex with Cpd637 ((R)-4-methyl-6-(1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-4-methyl-6-[1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
3ZU5
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU3
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (MR, cleaved Histag) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU2
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (SIRAS) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, SODIUM ION | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
6CXC
 
 | | 3.9A Cryo-EM structure of murine antibody bound at a novel epitope of respiratory syncytial virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, Envelope glycoprotein chimera, ... | | Authors: | Xie, Q, Wang, Z, Chen, X, Ni, F, Ma, J, Wang, Q. | | Deposit date: | 2018-04-02 | | Release date: | 2019-07-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein.
Plos One, 14, 2019
|
|
6LRM
 
 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
5UV6
 
 | | Crystal structure of human Opioid Binding Protein/Cell Adhesion Molecule Like (OPCML) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Opioid-binding protein/cell adhesion molecule, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Birtley, J.R, Stern, L.J, Gabra, H, Zanini, E. | | Deposit date: | 2017-02-19 | | Release date: | 2018-03-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65002 Å) | | Cite: | Inactivating mutations and X-ray crystal structure of the tumor suppressor OPCML reveal cancer-associated functions.
Nat Commun, 10, 2019
|
|
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
6VY2
 
 | |
5VLR
 
 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A TRIFLUORO-ETHYL-PYRAZOL-PYROLOTRIAZINE INHIBITOR | | Descriptor: | 4-acetyl-1-(3-{4-amino-5-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)-3,3-dimethylpiperazin-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Sack, J.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Potent, Selective, and Efficacious Phosphatidylinositol 3-Kinase delta (PI3K delta ) Inhibitor for the Treatment of Immunological Disorders.
J. Med. Chem., 60, 2017
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
5E8Y
 
 | |
5E8W
 
 | |
5E8T
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) | | Descriptor: | GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E90
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F, S280T,Y282F,S287N,A350C,L352F) IN COMPLEX WITH 3-AMINO-6- [4-(2-HYDROXYETHYL)PHENYL]-N-[4-(MORPHOLIN-4-YL)PYRIDIN-3-YL]PYRAZINE-2-CARBOXAMIDE | | Descriptor: | 3-amino-6-[4-(2-hydroxyethyl)phenyl]-N-[4-(morpholin-4-yl)pyridin-3-yl]pyrazine-2-carboxamide, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E8U
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F,S280T, Y282F,S287N,A350C,L352F) | | Descriptor: | GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E91
 
 | |
5E92
 
 | | TGF-BETA RECEPTOR TYPE 2 KINASE DOMAIN (E431A,R433A,E485A,K488A,R493A,R495A) IN COMPLEX WITH AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TGF-beta receptor type-2 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5E8S
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (WT) | | Descriptor: | TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
