8DN3
 
 | |
8DN2
 
 | |
8DN5
 
 | |
4EEZ
 
 | | Crystal Structure of Lactococcus lactis Alcohol Dehydrogenase variant RE1 | | Descriptor: | Alcohol dehydrogenase 1, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Liu, X, Bastian, S, Snow, C.D, Brustad, E.M, Saleski, T, Xu, J.H, Meinhold, P, Arnold, F.H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol.
J.Biotechnol., 164, 2012
|
|
4EEX
 
 | | Crystal Structure of Lactococcus lactis Alcohol Dehydrogenase | | Descriptor: | Alcohol dehydrogenase 1, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Liu, X, Bastian, S, Snow, C.D, Brustad, E.M, Saleski, T, Xu, J.H, Meinhold, P, Arnold, F.H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol.
J.Biotechnol., 164, 2012
|
|
6EXV
 
 | | Structure of mammalian RNA polymerase II elongation complex inhibited by Alpha-amanitin | | Descriptor: | AMATOXIN, DNA (25-MER), DNA (36-MER), ... | | Authors: | Liu, X, Farnung, L, Wigge, C, Cramer, P. | | Deposit date: | 2017-11-09 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by alpha-amanitin.
J. Biol. Chem., 293, 2018
|
|
5GWM
 
 | |
4YR8
 
 | | Crystal structure of JNK in complex with a regulator protein | | Descriptor: | CHLORIDE ION, Dual specificity protein phosphatase 16, Mitogen-activated protein kinase 8 | | Authors: | Liu, X, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2015-03-14 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A conserved motif in JNK/p38-specific MAPK phosphatases as a determinant for JNK1 recognition and inactivation.
Nat Commun, 7, 2016
|
|
3RZD
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3RZO
 
 | | RNA Polymerase II Initiation Complex with a 4-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-12 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1R
 
 | | RNA Polymerase II Initiation Complex with a 5-nt 3'-deoxy RNA soaked with GTP | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S16
 
 | | RNA Polymerase II Initiation Complex with an 8-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.241 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S14
 
 | | RNA Polymerase II Initiation Complex with a 6-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1N
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA (variant 2) | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*GP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-15 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S2D
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA containing a 5Br-U | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*AP*CP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1Q
 
 | | RNA Polymerase II Initiation Complex with a 5-nt 3'-deoxy RNA soaked with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S15
 
 | | RNA Polymerase II Initiation Complex with a 7-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S2H
 
 | | RNA Polymerase II Initiation Complex with a 6-nt RNA containing a 2[prime]-iodo ATP | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S17
 
 | | RNA Polymerase II Initiation Complex with a 9-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1M
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA (variant 1) | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*GP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-15 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3TEF
 
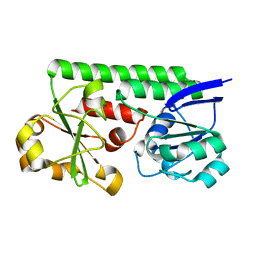 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
3BIY
 
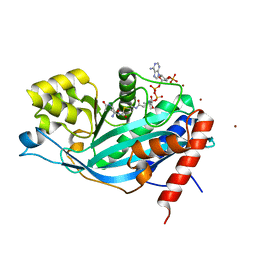 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
3C20
 
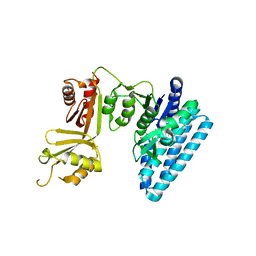 | |
3C1N
 
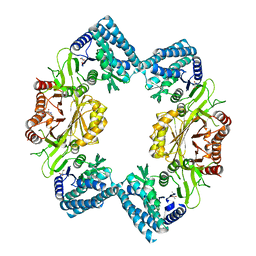 | |
3C1M
 
 | |
