7Q6Q
 
 | | Keap1 compound complex | | Descriptor: | (5S,8R)-N,N-dimethyl-8-[[(2S)-1-[4-(methylamino)-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(12),13,15-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q5H
 
 | | Keap1 compound complex | | Descriptor: | (3S,5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-3,7,11-tris(oxidanylidene)-10-oxa-3$l^{4}-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q96
 
 | | Keap1 compound complex | | Descriptor: | 4-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q6S
 
 | | Keap1 compound complex | | Descriptor: | (5S,8R)-16-(2,1,3-benzoxadiazol-4-yl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q8R
 
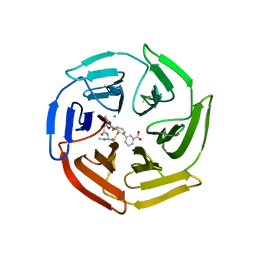 | | Keap1 compound complex | | Descriptor: | 3-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
6OVJ
 
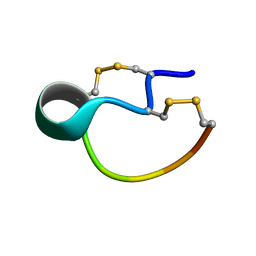 | |
6PPC
 
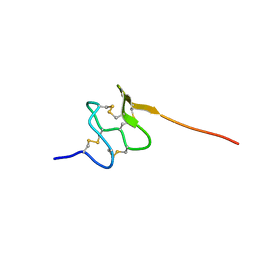 | | Solution structure of conotoxin MiXXVIIA | | Descriptor: | Conopeptide phi-MiXXVIIA | | Authors: | Daly, N.L, Dekan, Z, Jin, A.H, Alewood, P.F. | | Deposit date: | 2019-07-06 | | Release date: | 2019-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conotoxin phi-MiXXVIIA from the Superfamily G2 Employs a Novel Cysteine Framework that Mimics Granulin and Displays Anti-Apoptotic Activity.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7N0T
 
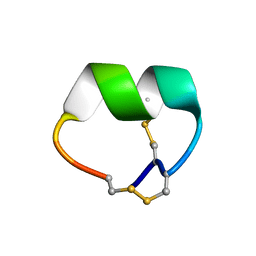 | | NMR structure of EpI[Y(SO)315Y]-OH | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N25
 
 | | NMR structure of EpI-OH | | Descriptor: | Alpha-conotoxin EpI-OH | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N26
 
 | | NMR structure of EpI-[Y(SO3)15Y]-NH2 | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N22
 
 | | NMR structure of AnIB[Y(SO3)16Y]-NH2 | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N24
 
 | | NMR structure of native EpI | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N20
 
 | | NMR structure of native AnIB | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N21
 
 | | NMR structure of AnIB-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N1Z
 
 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N23
 
 | | NMR structure of AnIB[Y(SO3)16Y]-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7O0I
 
 | | Vibrio vulnificus stressosome | | Descriptor: | Anti-anti-sigma factor, RsbS, negative regulator of sigma-B | | Authors: | Kaltwasser, S, Heinz, V, Madej, M.G, Pane-Farre, J, Ziegler, C. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The Vibrio vulnificus stressosome is an oxygen-sensor involved in regulating iron metabolism
Commun Biol, 2022
|
|
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
5T3M
 
 | |
5T4R
 
 | |
2MT7
 
 | |
2MPQ
 
 | |
2MXM
 
 | |
2N7F
 
 | | NMR solution structure of muO-conotoxin MfVIA | | Descriptor: | muO-conotoxin MfVIA | | Authors: | Schroeder, C.I, Mobli, M. | | Deposit date: | 2015-09-09 | | Release date: | 2016-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of a mu O-Conotoxin Analogue with Improved Lipid Membrane Interactions and Potency for the Analgesic Sodium Channel NaV1.8.
J.Biol.Chem., 291, 2016
|
|
2KRA
 
 | |
