8AI4
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI5
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI1
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI2
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI6
 
 | | Crystal structure of radical SAM epimerase EpeE D210A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and persulfurated cysteine bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI3
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-methionine bound | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Chavas, L.M.G, Legrand, P, Polsinelli, I, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPC
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2683593 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZP9
 
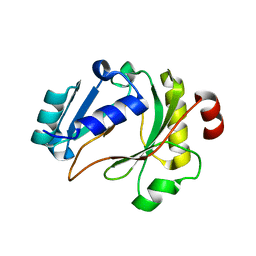 | |
6ZPB
 
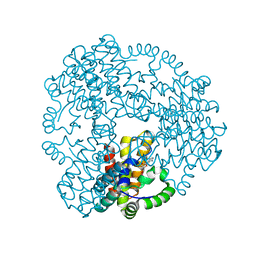 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and two catalytic Co2+ ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, COBALT (II) ION, DatZ | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72097385 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZUF
 
 | | Urea-based Foldamer Inhibitor chimera C2 in complex with ASF1 Histone chaperone | | Descriptor: | C2 foldamer/peptide hybrid inhibitor of histone chaperone ASF1, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Mbianda, J, Perrin, E.M, Guerois, R, Legrand, P, Traore, S, Douat, C, Guichard, G, Ochsenbein, F. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity.
Sci Adv, 7, 2021
|
|
7AWQ
 
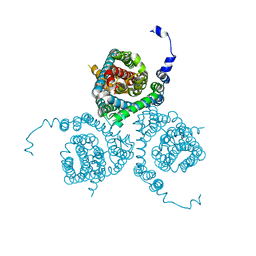 | | Structure of the thermostabilized EAAT1 cryst-E386Q mutant in complex with L-ASP, sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWM
 
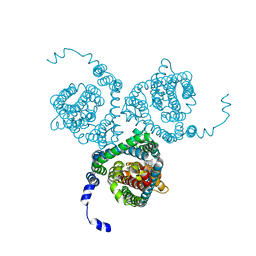 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP, three sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWN
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with rubidium and barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWL
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWP
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with rubidium and barium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7ANQ
 
 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
7BDX
 
 | | Armadillo domain of HSF2BP in complex with BRCA2 peptide | | Descriptor: | Breast cancer type 2 susceptibility protein, Heat shock factor 2-binding protein, MAGNESIUM ION | | Authors: | Le Du, M.H, Zinn-Justin, S, Ghouil, R, Miron, S, Legrand, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BRCA2 binding through a cryptic repeated motif to HSF2BP oligomers does not impact meiotic recombination.
Nat Commun, 12, 2021
|
|
8AA9
 
 | |
8B8D
 
 | | multimerization domain of Gaboon Viper Virus 1 | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bouhris, J.M, Horie, M, Tomonaga, K, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
8B8A
 
 | | Multimerization domain of borna disease virus 1 phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bourhis, J.M, Chenavier, F, Freslon, L, Kawasaki, J, Horie, M, Tomonaga, K, Bachiri, K, Coyaud, E, Gonzalez-Dunia, D, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
8AUC
 
 | | Structure of peptidoglycan hydrolase Cg1735 from Corynebacterium glutamicum, trigonal crystal form | | Descriptor: | Cell wall-associated hydrolases (Invasion-associated proteins), TETRACHLOROPLATINATE(II) | | Authors: | Gaday, Q, Wehenkel, A.M, Alzari, P.M, Legrand, P. | | Deposit date: | 2022-08-25 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | FtsEX-independent control of RipA-mediated cell separation in Corynebacteriales.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8BV1
 
 | | Peptide inhibitor P4 in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, P4 peptide inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.834 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ3
 
 | | Urea-based foldamer inhibitor c3u_7 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_7 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ2
 
 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
