7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
5TEC
 
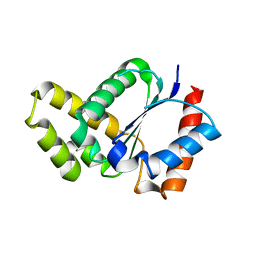 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | Descriptor: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | Authors: | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2016-09-20 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VJJ
 
 | | Crystal structure of the flax-rust effector AvrP | | Descriptor: | Avirulence protein AvrP123, ZINC ION | | Authors: | Zhang, X, Ericsson, D.J, Williams, S.J, Kobe, B. | | Deposit date: | 2017-04-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the Melampsora lini effector AvrP reveals insights into a possible nuclear function and recognition by the flax disease resistance protein P.
Mol. Plant Pathol., 19, 2018
|
|
6VZB
 
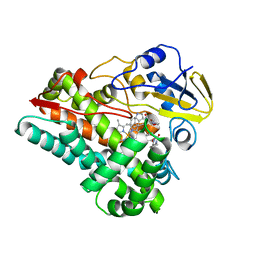 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VZA
 
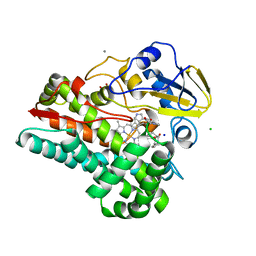 | | Crystal structure of cytochrome P450 NasF5053 Q65I-A86G mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VXV
 
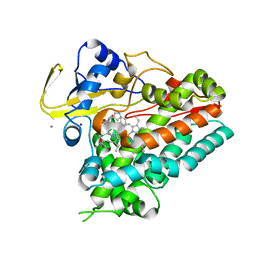 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6W0S
 
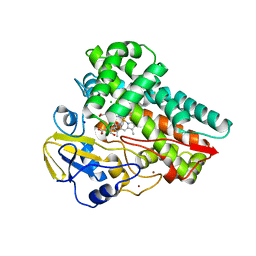 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
5UZB
 
 | | Cryo-EM structure of the MAL TIR domain filament | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Ve, T, Vajjhala, P.R, Hedger, A, Croll, T, DiMaio, F, Horsefield, S, Yu, X, Lavrencic, P, Hassan, Z, Morgan, G.P, Mansell, A, Mobli, M, O'Carrol, A, Chauvin, B, Gambin, Y, Sierecki, E, Landsberg, M.J, Stacey, K.J, Egelman, E.H, Kobe, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-07-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3L3Q
 
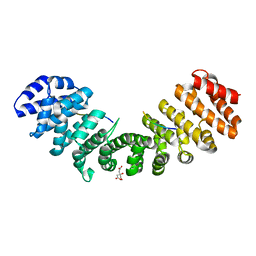 | | Mouse importin alpha-pepTM NLS peptide complex | | Descriptor: | CITRATE ANION, Importin subunit alpha-2, pepTM | | Authors: | Takeda, A.A.S, Kobe, B, Fontes, M.R.M. | | Deposit date: | 2009-12-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening.
J.Biol.Chem., 285, 2010
|
|
3ND2
 
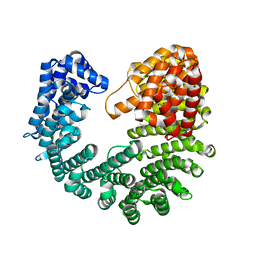 | | Structure of Yeast Importin-beta (Kap95p) | | Descriptor: | Importin subunit beta-1 | | Authors: | Forwood, J.K, Kobe, B. | | Deposit date: | 2010-06-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Quantitative Structural Analysis of Importin-beta Flexibility: Paradigm for Solenoid Protein Structures
Structure, 18, 2010
|
|
3OZI
 
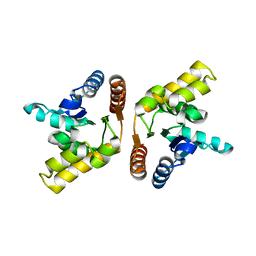 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | Descriptor: | COBALT (II) ION, L6tr | | Authors: | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2010-09-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
5D90
 
 | |
5D8C
 
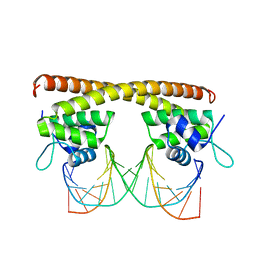 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to promoter DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*AP*AP*CP*TP*CP*TP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*TP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3'), MerR family regulator protein | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
5E01
 
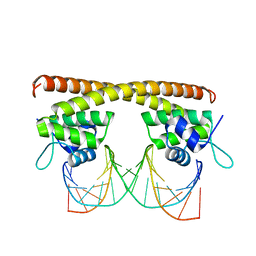 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to palyndromic promoter DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3', Uncharacterized HTH-type transcriptional regulator HI_0186 | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-09-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
2OPC
 
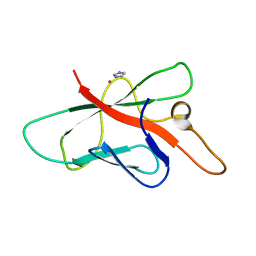 | | Structure of Melampsora lini avirulence protein, AvrL567-A | | Descriptor: | AvrL567-A, COBALT (II) ION, IMIDAZOLE | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-01-29 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
7JJA
 
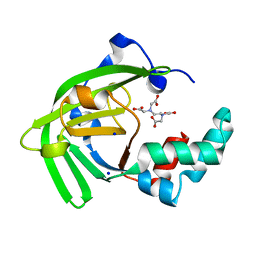 | | Crystal structure of the ZinT-like domain of Streptococcus pneumoniae AdcA in the apo form | | Descriptor: | SODIUM ION, Zinc-binding lipoprotein AdcA, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7JJ9
 
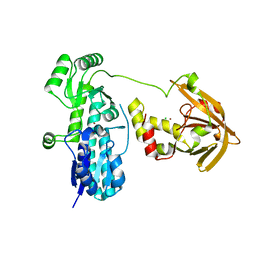 | | Crystal structure of Zn(II)-bound AdcA from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7JJ8
 
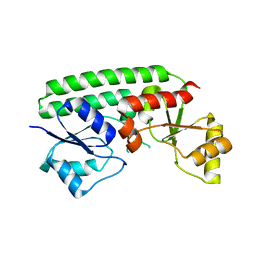 | | Crystal structure of the Zn(II)-bound ZnuA-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7JJB
 
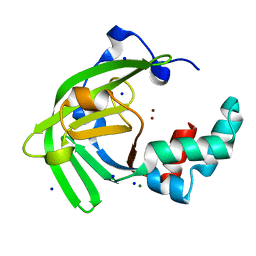 | | Crystal structure of Zn(II)-bound ZinT-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | MAGNESIUM ION, SODIUM ION, ZINC ION, ... | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7L6W
 
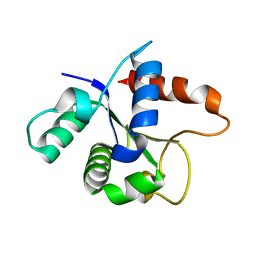 | | SFX structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7L5S
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7L5I
 
 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
6N8A
 
 | | Crystal structure of selenomethionine-containing AcaB from uropathogenic E. coli | | Descriptor: | CHLORIDE ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-28 | | Release date: | 2020-07-15 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
6N8B
 
 | | Crystal structure of transcription regulator AcaB from uropathogenic E. coli | | Descriptor: | CALCIUM ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
