6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
1CWP
 
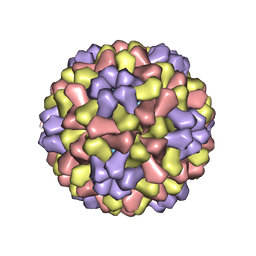 | | STRUCTURES OF THE NATIVE AND SWOLLEN FORMS OF COWPEA CHLOROTIC MOTTLE VIRUS DETERMINED BY X-RAY CRYSTALLOGRAPHY AND CRYO-ELECTRON MICROSCOPY | | Descriptor: | Coat protein, RNA (5'-R(*AP*U)-3'), RNA (5'-R(*AP*UP*AP*U)-3') | | Authors: | Speir, J.A, Johnson, J.E, Munshi, S, Wang, G, Timothy, S, Baker, T.S. | | Deposit date: | 1995-05-22 | | Release date: | 1995-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy.
Structure, 3, 1995
|
|
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1A1R
 
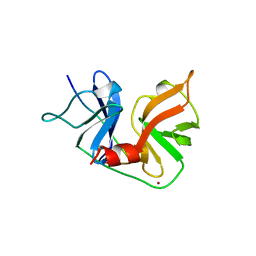 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
1CPT
 
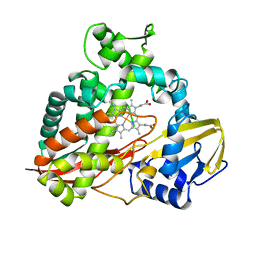 | | CRYSTAL STRUCTURE AND REFINEMENT OF CYTOCHROME P450-TERP AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME P450-TERP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasemann, C.A, Ravichandran, K.G, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and refinement of cytochrome P450terp at 2.3 A resolution.
J.Mol.Biol., 236, 1994
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4US3
 
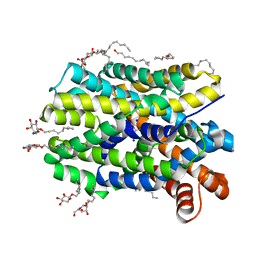 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
8GHR
 
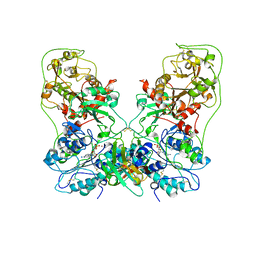 | | Structure of human ENPP1 in complex with variable heavy domain VH27.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Carozza, J.A, Wang, H, Solomon, P.E, Wells, J.A, Li, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of VH domains that allosterically inhibit ENPP1.
Nat.Chem.Biol., 20, 2024
|
|
4US4
 
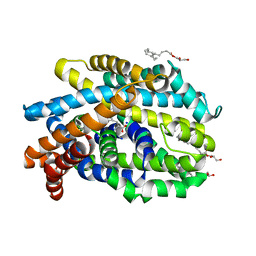 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
1DKT
 
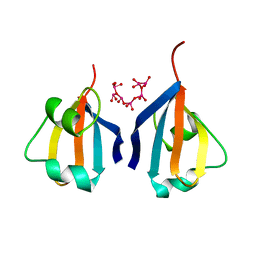 | | CKSHS1: HUMAN CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1 COMPLEX WITH METAVANADATE | | Descriptor: | CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1, META VANADATE | | Authors: | Bourne, Y, Arvai, A.S, Tainer, J.A. | | Deposit date: | 1995-11-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain.
J.Mol.Biol., 249, 1995
|
|
1DF1
 
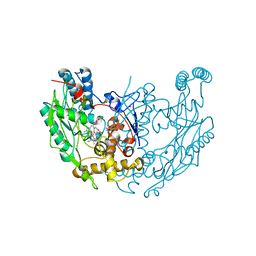 | | MURINE INOSOXY DIMER WITH ISOTHIOUREA BOUND IN THE ACTIVE SITE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYLISOTHIOUREA, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Rosenfeld, R.J, Arvai, A.S, Ghosh, D.K, Ghosh, S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-16 | | Release date: | 1999-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
EMBO J., 18, 1999
|
|
7BGS
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BNX
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8SF8
 
 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
7N1S
 
 | | Crystal Structure Analysis of Xac Nucleotide Pyrophosphatase/Phosphodiesterase | | Descriptor: | Phosphodiesterase-nucleotide pyrophosphatase, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Fernandez, D, Li, L, Brown, J.A, Carozza, J.A. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ENPP1's regulation of extracellular cGAMP is a ubiquitous mechanism of attenuating STING signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U9U
 
 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | (3R)-3-(5,6-dioxo-1,4,5,6-tetrahydropyrazin-2-yl)-2,3-dihydro-1,4-benzoxathiine-7-carbonitrile, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7U9S
 
 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | 5-{2-[4-(trifluoromethyl)phenyl]ethyl}-1,4-dihydropyrazine-2,3-dione, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7USL
 
 | | Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
7USM
 
 | | Integrin alphaM/beta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
7UMO
 
 | | Structure of Unc119-inhibitor complex. | | Descriptor: | (3s,5s,7s)-N-(4,5-dichloropyridin-2-yl)adamantane-1-carboxamide, GLYCEROL, Protein unc-119 homolog A | | Authors: | Srivastava, D, Sebag, J.A, Artemyev, N.O. | | Deposit date: | 2022-04-07 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin sensitization by small molecules enhancing GLUT4 translocation.
Cell Chem Biol, 30, 2023
|
|
2ACL
 
 | | Liver X-Receptor alpha Ligand Binding Domain with SB313987 | | Descriptor: | 1-BENZYL-3-(4-METHOXYPHENYLAMINO)-4-PHENYLPYRROLE-2,5-DIONE, Oxysterols receptor LXR-alpha, RETINOIC ACID, ... | | Authors: | Jaye, M.C, Krawiec, J.A, Campobasso, N, Smallwood, A, Qiu, C, Lu, Q, Kerrigan, J.J. | | Deposit date: | 2005-07-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of substituted maleimides as liver x receptor agonists and determination of a ligand-bound crystal structure.
J.Med.Chem., 48, 2005
|
|
1BKH
 
 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BQG
 
 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
