5OA0
 
 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, W164F) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, STRYCHNINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
5O87
 
 | | Crystal structure of wild type Aplysia californica AChBP in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
5OBH
 
 | | Crystal structure of glycine binding protein in complex with bicuculline | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dawson, A, Hunter, W.N, Jones, M. | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Rationale for N-Methylbicuculline Acting as a Promiscuous Competitive Antagonist of Inhibitory Pentameric Ligand-Gated Ion Channels.
Chembiochem, 21, 2020
|
|
5OPZ
 
 | | Crystal structure of Serratia marcescens L-Ala D-Glu endopeptidase ChiX | | Descriptor: | CHLORIDE ION, ChiX, ZINC ION | | Authors: | Owen, R.A, Fyfe, P.K, Lodge, A, Biboy, J, Vollmer, W, Hunter, W.N, Sargent, F. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens.
Biochem. J., 475, 2018
|
|
5OQ1
 
 | | Crystal structure of Serratia marcescens ChiX (used as MR model for superior PDB 5OPZ) | | Descriptor: | CHLORIDE ION, ChiX, ZINC ION | | Authors: | Owen, R.A, Fyfe, P.K, Lodge, A, Biboy, J, Vollmer, W, Hunter, W.N, Sargent, F. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens.
Biochem. J., 475, 2018
|
|
110D
 
 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*GP*CP*CP*G)-3') | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
112D
 
 | |
114D
 
 | | INOSINE-ADENINE BASE PAIRS IN A B-DNA DUPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*IP*AP*AP*TP*TP*AP*GP*CP*G)-3') | | Authors: | Corfield, P.W.R, Hunter, W.N, Brown, T, Robinson, P, Kennard, O. | | Deposit date: | 1993-01-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inosine.adenine base pairs in a B-DNA duplex.
Nucleic Acids Res., 15, 1987
|
|
119D
 
 | |
133D
 
 | | THE CRYSTAL STRUCTURE OF N4-METHYLCYTOSINE.GUANOSIN BASE-PAIRS IN THE SYNTHETIC HEXANUCLEOTIDE D(CGCGM(4)CG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C34)P*G)-3') | | Authors: | Cervi, A.R, Guy, A, Leonard, G.A, Teoule, R, Hunter, W.N. | | Deposit date: | 1993-07-29 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of N4-methylcytosine.guanosine base-pairs in the synthetic hexanucleotide d(CGCGm4CG).
Nucleic Acids Res., 21, 1993
|
|
178D
 
 | | CRYSTAL STRUCTURE OF A DNA DUPLEX CONTAINING 8-HYDROXYDEOXYGUANINE.ADENINE BASE-PAIRS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*(8OG)P*GP*CP*G)-3') | | Authors: | McAuley-Hecht, K.E, Leonard, G.A, Gibson, N.J, Thomson, J.B, Watson, W.P, Hunter, W.N, Brown, T. | | Deposit date: | 1994-05-19 | | Release date: | 1994-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs.
Biochemistry, 33, 1994
|
|
157D
 
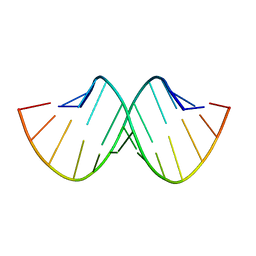 | | CRYSTAL AND MOLECULAR STRUCTURE OF R(CGCGAAUUAGCG): AN RNA DUPLEX CONTAINING TWO G(ANTI).A(ANTI) BASE-PAIRS | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Ebel, S, Lough, D.M, Brown, T, Hunter, W.N. | | Deposit date: | 1994-02-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti).A(anti) base pairs.
Structure, 2, 1994
|
|
184D
 
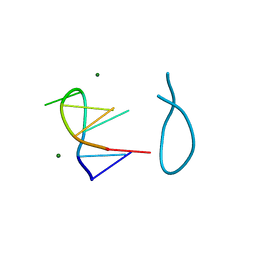 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
198D
 
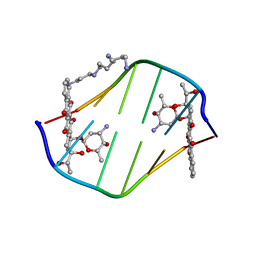 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
150D
 
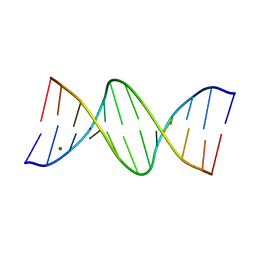 | | GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDA)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Gibson, N.J, Brown, T, Watson, W.P, Hunter, W.N. | | Deposit date: | 1993-12-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG).
Biochemistry, 33, 1994
|
|
6QKK
 
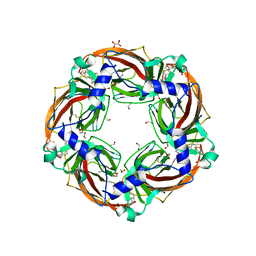 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]benzamide, ... | | Authors: | Davis, S, Bueno, R.V, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
6QQP
 
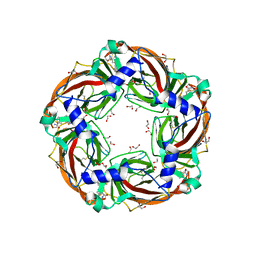 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (2) | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]pyridine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bueno, R.V, Davis, S, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
6QQO
 
 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]pyridine-3-carboxamide, ... | | Authors: | Bueno, R.V, Davis, S, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
6T9R
 
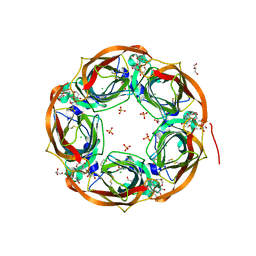 | | Aplysia californica AChBP in complex with a cytisine derivative | | Descriptor: | (1~{R},9~{S})-5-(3-oxidanylpropyl)-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine binding protein, ... | | Authors: | Davis, S, Hunter, W.N. | | Deposit date: | 2019-10-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The thermodynamic profile and molecular interactions of a C(9)-cytisine derivative-binding acetylcholine-binding protein from Aplysia californica.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2HKE
 
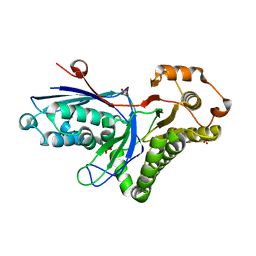 | | Mevalonate diphosphate decarboxylase from Trypanosoma brucei | | Descriptor: | Diphosphomevalonate decarboxylase, putative, SULFATE ION | | Authors: | Byres, E, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2006-07-04 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Trypanosoma brucei and Staphylococcus aureus Mevalonate Diphosphate Decarboxylase Inform on the Determinants of Specificity and Reactivity
J.Mol.Biol., 371, 2007
|
|
1H47
 
 | | Structures of MECP synthase in complex with (i) CMP and (ii) CMP and product | | Descriptor: | 2C-METHYL-D-ERYTHRITOL-2,4-CYCLODIPHOSPHATE SYNTHASE, CYTIDINE-5'-MONOPHOSPHATE, GERANYL DIPHOSPHATE, ... | | Authors: | Kemp, L.E, Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2003-02-24 | | Release date: | 2004-10-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Identification of Isoprenoids that Bind in the Intersubunit Cavity of Escherichia Coli 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase by Complementary Biophysical Methods
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W8D
 
 | | Binary structure of human DECR. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, GLYCEROL, ... | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
1W73
 
 | | Binary structure of human DECR solved by SeMet SAD. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
1W77
 
 | | 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD) from Arabidopsis thaliana | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CADMIUM ION, COPPER (II) ION, ... | | Authors: | Gabrielsen, M, Kaiser, J, Rohdich, F, Eisenreich, W, Bacher, A, Bond, C.S, Hunter, W.N. | | Deposit date: | 2004-08-30 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Plant 2C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase Exhibits a Distinct Quaternary Structure Compared to Bacterial Homologues and a Possible Role in Feedback Regulation for Cytidine Monophosphate.
FEBS J., 273, 2006
|
|
1W6U
 
 | | Structure of human DECR ternary complex | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, HEXANOYL-COENZYME A, ... | | Authors: | Alphey, M.S, Byres, E, Li, D, Hunter, W.N. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
