3SP4
 
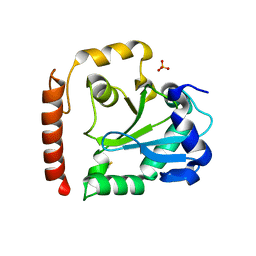 | | Crystal structure of aprataxin ortholog Hnt3 from Schizosaccharomyces pombe | | 分子名称: | Aprataxin-like protein, SULFATE ION, ZINC ION | | 著者 | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | 登録日 | 2011-07-01 | | 公開日 | 2011-10-12 | | 最終更新日 | 2013-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SPL
 
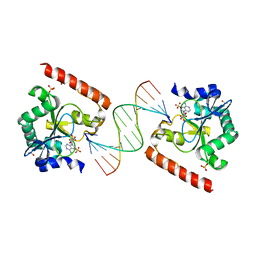 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA and AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), ... | | 著者 | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | 登録日 | 2011-07-02 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SPD
 
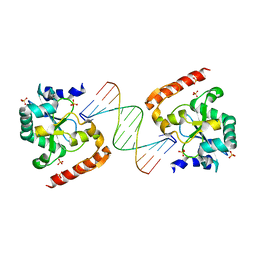 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA | | 分子名称: | Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*AP*C)-3'), ... | | 著者 | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | 登録日 | 2011-07-01 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.912 Å) | | 主引用文献 | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3BUK
 
 | | Crystal Structure of the Neurotrophin-3 and p75NTR Symmetrical Complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neurotrophin-3, Tumor necrosis factor receptor superfamily member 16 | | 著者 | Jiang, T, Gong, Y, Cao, P, Yu, H.J. | | 登録日 | 2008-01-02 | | 公開日 | 2008-07-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex.
Nature, 454, 2008
|
|
2JR8
 
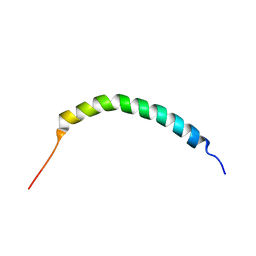 | | Solution structure of Manduca sexta moricin | | 分子名称: | Antimicrobial peptide moricin | | 著者 | Gong, Y, Dai, H, Rayaprolu, S, Huang, R, Prakash, O, Jiang, H. | | 登録日 | 2007-06-21 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure, antibacterial activity, and expression profile of Manduca sexta moricin.
J.Pept.Sci., 14, 2008
|
|
7CHU
 
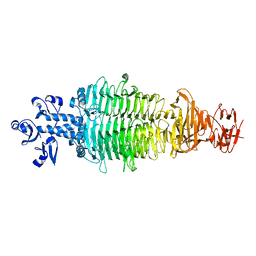 | | Geobacillus virus E2 - ORF18 | | 分子名称: | Putative pectin lyase | | 著者 | Gong, Y. | | 登録日 | 2020-07-06 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Structural and functional characterization of the deep-sea thermophilic bacteriophage GVE2 tailspike protein.
Int.J.Biol.Macromol., 164, 2020
|
|
1MIT
 
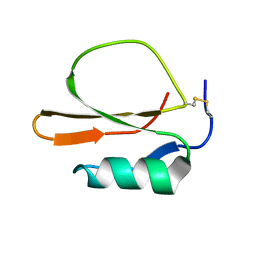 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | 分子名称: | TRYPSIN INHIBITOR V | | 著者 | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | 登録日 | 1995-10-26 | | 公開日 | 1996-04-03 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
5K07
 
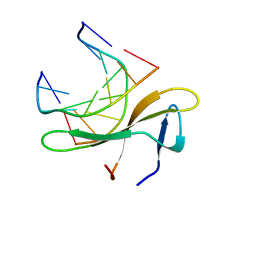 | | Crystal structure of CREN7-DSDNA (GTAATTGC) complex | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*GP*C)-3') | | 著者 | Zhang, Z.F, Gong, Y. | | 登録日 | 2016-05-17 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea
PLoS ONE, 11, 2016
|
|
5K17
 
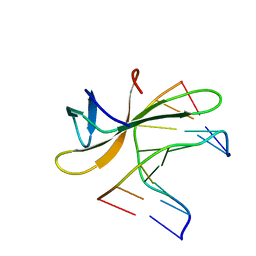 | | Crystal structure of CREN7-DSDNA (GTGATCGC) complex | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3') | | 著者 | Zhang, Z.F, Gong, Y. | | 登録日 | 2016-05-17 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea
PLoS ONE, 11, 2016
|
|
1HYM
 
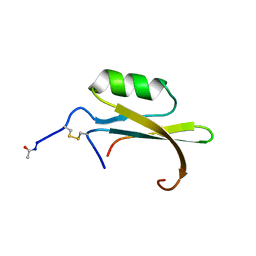 | | HYDROLYZED TRYPSIN INHIBITOR (CMTI-V, MINIMIZED AVERAGE NMR STRUCTURE) | | 分子名称: | HYDROLYZED CUCURBITA MAXIMA TRYPSIN INHIBITOR V | | 著者 | Cai, M, Gong, Y, Prakash, O, Krishnamoorthi, R. | | 登録日 | 1995-06-12 | | 公開日 | 1995-09-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Reactive-site hydrolyzed Cucurbita maxima trypsin inhibitor-V: function, thermodynamic stability, and NMR solution structure.
Biochemistry, 34, 1995
|
|
8WKH
 
 | | Crystal structure of group 13 allergen from Blomia tropicalis | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Fatty acid-binding protein | | 著者 | Zhu, K.L, Gong, Y, Cui, Y.B. | | 登録日 | 2023-09-27 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Immunobiological properties and structure analysis of group 13 allergen from Blomia tropicalis and its IgE-mediated cross-reactivity.
Int.J.Biol.Macromol., 254, 2023
|
|
6UI4
 
 | | Crystal structure of phenamacril-bound F. graminearum myosin I | | 分子名称: | Calmodulin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Zhou, Y, Zhou, X.E, Gong, Y, Zhu, Y, Xu, H.E, Zhou, M, Melcher, K, Zhang, F. | | 登録日 | 2019-09-30 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis of Fusarium myosin I inhibition by phenamacril.
Plos Pathog., 16, 2020
|
|
3EWT
 
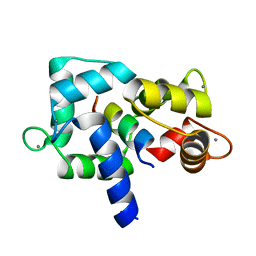 | | Crystal Structure of calmodulin complexed with a peptide | | 分子名称: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 6 | | 著者 | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | 登録日 | 2008-10-16 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3EWV
 
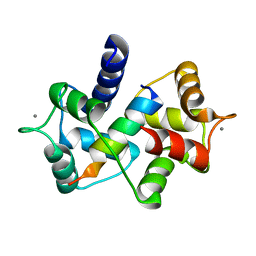 | | Crystal Structure of calmodulin complexed with a peptide | | 分子名称: | CALCIUM ION, Calmodulin, Tumor necrosis factor receptor superfamily member 16 | | 著者 | Jiang, T, Cao, P, Gong, Y, Yu, H.J, Gui, W.J, Zhang, W.T. | | 登録日 | 2008-10-16 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into the mechanism of calmodulin binding to death receptors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WVZ
 
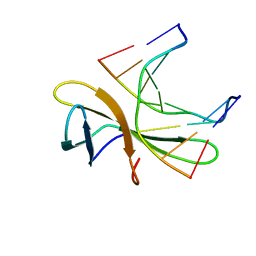 | | The crystal structure of Cren7 mutant L28F in complex with dsDNA | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | 著者 | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | 登録日 | 2016-12-30 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVY
 
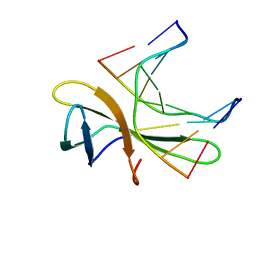 | | The crystal structure of Cren7 mutant L28V in complex with dsDNA | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | 著者 | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | 登録日 | 2016-12-29 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVW
 
 | | The crystal structure of Cren7 mutant L28A in complex with dsDNA | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | 著者 | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | 登録日 | 2016-12-29 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WWC
 
 | | The crystal structure of Cren7 mutant L28M in complex with dsDNA | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | 著者 | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | 登録日 | 2016-12-31 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
1TIN
 
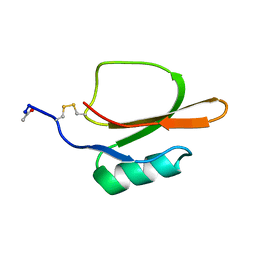 | |
1Z9M
 
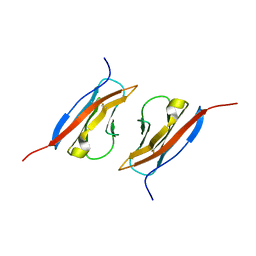 | | Crystal Structure of Nectin-like molecule-1 protein Domain 1 | | 分子名称: | GAPA225 | | 著者 | Dong, X, Xu, F, Gong, Y, Gao, J, Lin, P, Chen, T, Peng, Y, Qiang, B, Yuan, J, Peng, X, Rao, Z. | | 登録日 | 2005-04-03 | | 公開日 | 2006-02-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the V Domain of Human Nectin-like Molecule-1/Syncam3/Tsll1/Igsf4b, a Neural Tissue-specific Immunoglobulin-like Cell-Cell Adhesion Molecule
J.Biol.Chem., 281, 2006
|
|
5HW4
 
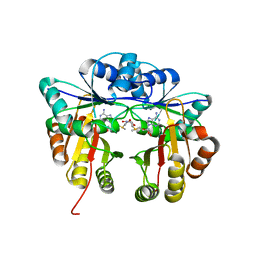 | | Crystal structure of Escherichia coli 16S rRNA methyltransferase RsmI in complex with AdoMet | | 分子名称: | Ribosomal RNA small subunit methyltransferase I, S-ADENOSYLMETHIONINE | | 著者 | Zhao, M, Zhang, H, Dong, Y, Gong, Y. | | 登録日 | 2016-01-28 | | 公開日 | 2016-10-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.211 Å) | | 主引用文献 | Structural Insights into the Methylation of C1402 in 16S rRNA by Methyltransferase RsmI
Plos One, 11, 2016
|
|
4R56
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA(GTGATCAC) complex | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | 著者 | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | 登録日 | 2014-08-20 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
4R55
 
 | | The crystal structure of a Cren7 mutant protein GR and dsDNA complex | | 分子名称: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | 著者 | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | 登録日 | 2014-08-20 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
4RXS
 
 | | The structure of GTP-dTTP-bound SAMHD1 | | 分子名称: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | 登録日 | 2014-12-11 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RXR
 
 | | The structure of GTP-dCTP-bound SAMHD1 | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | 登録日 | 2014-12-11 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.117 Å) | | 主引用文献 | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
