4MQT
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, ... | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
4MXQ
 
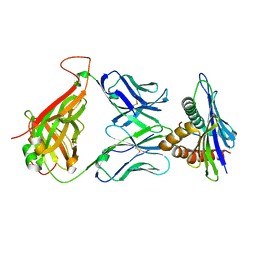 | | 42F3 TCR pCPC5/H-2Ld Complex | | Descriptor: | 42F3 alpha VmVh chimera, 42F3 beta VmVh chimera, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-09-26 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4MVB
 
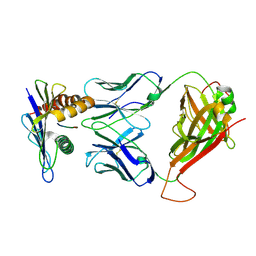 | | 42F3 pCPB7/H-2Ld Complex | | Descriptor: | 42F3 alpha VmCh, 42F3 beta VmCh, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-09-23 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (3.088 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4N5E
 
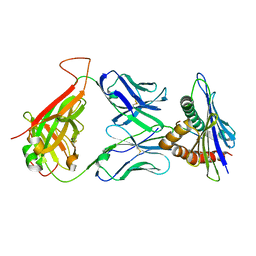 | | 42F3 TCR pCPA12/H-2Ld complex | | Descriptor: | 42F3 alpha VmCh, 42F3 beta VmCh, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-10-09 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (3.059 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4N0C
 
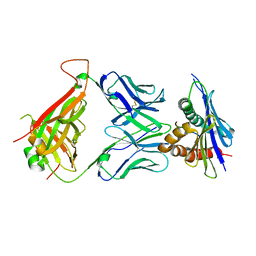 | | 42F3 TCR pCPE3/H-2Ld complex | | Descriptor: | 42F3 VmCh alpha, 42F3 VmCh beta, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-10-01 | | Release date: | 2015-08-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity.
Nat. Immunol., 17, 2016
|
|
4NHU
 
 | | The M33 TCR p3M33l/H-2 Ld Complex | | Descriptor: | 2C m33 alpha VmCh chimera, 2C m33 beta VmCh chimera, H-2 class I histocompatibility antigen, ... | | Authors: | Birnbaum, M.E, Adams, J.J, Garcia, K.C. | | Deposit date: | 2013-11-05 | | Release date: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interrogating TCR Signal Strength and Cross-Reactivity by Yeast Display of pMHC
To be Published
|
|
4OFD
 
 | | Crystal Structure of mouse Neph1 D1-D2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of IRRE-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFI
 
 | | Crystal Structure of Duf (Kirre) D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of irre, isoform A, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OQW
 
 | | Crystal structure of mCardinal far-red fluorescent protein | | Descriptor: | Fluorescent protein FP480 | | Authors: | Burg, J.S, Chu, J, Lam, A.J, Lin, M.Z, Garcia, K.C. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
4OF3
 
 | | Crystal Structure of SYG-1 D1-D2, Glycosylated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFP
 
 | | Crystal Structure of SYG-2 D3-D4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-2 | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF8
 
 | | Crystal Structure of Rst D1-D2 | | Descriptor: | GLYCEROL, Irregular chiasm C-roughest protein, SODIUM ION | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF7
 
 | | Crystal Structure of SYG-1 D1, Crystal Form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFY
 
 | | Crystal Structure of the Complex of SYG-1 D1-D2 and SYG-2 D1-D4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHYL MERCURY ION, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF0
 
 | | Crystal Structure of SYG-1 D1-D2, refolded | | Descriptor: | Protein SYG-1, isoform b | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OFK
 
 | | Crystal Structure of SYG-2 D4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ozkan, E, Borek, D, Otwinowski, Z, Garcia, K.C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4OF6
 
 | | Crystal Structure of SYG-1 D1, Crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein SYG-1, isoform b, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4P2Q
 
 | | Crystal structure of the 5cc7 TCR in complex with 5c2/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5c2 peptide, 5cc7 T-cell receptor alpha chain, ... | | Authors: | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
4P2R
 
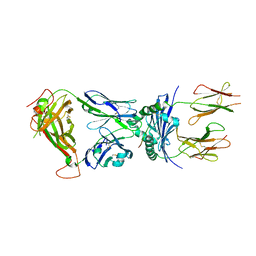 | | Crystal structure of the 5cc7 TCR in complex with 5c1/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5c1 peptide, 5cc7 T-cell receptor alpha chain, ... | | Authors: | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
4P2O
 
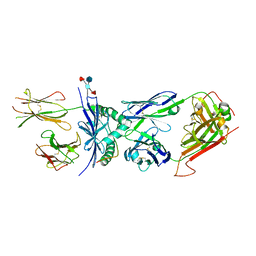 | | Crystal structure of the 2B4 TCR in complex with 2A/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2A peptide, 2B4 T-cell receptor alpha chain, ... | | Authors: | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
7JHJ
 
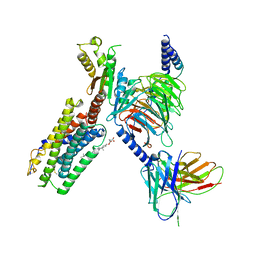 | | Structure of the Epstein-Barr virus GPCR BILF1 in complex with human Gi | | Descriptor: | Antibody fragment scFv16, BILF1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Tsutsumi, N, Qu, Q.H, Skiniotis, G, Garcia, K.C. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the constitutive activity and immunomodulatory properties of the Epstein-Barr virus-encoded G protein-coupled receptor BILF1.
Immunity, 54, 2021
|
|
1LDP
 
 | |
1ES0
 
 | |
7N1J
 
 | |
7N1K
 
 | |
