6AC8
 
 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
6ACX
 
 | | Crystal structure of Mycobacterium smegmatis Mfd in complex with ADP + Pi at 3.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Mycobacterium smegmatis Mfd, PHOSPHATE ION, ... | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
2IZP
 
 | | BipD - an invasion protein associated with the type-III secretion system of Burkholderia pseudomallei. | | Descriptor: | PUTATIVE MEMBRANE ANTIGEN | | Authors: | Erskine, P.T, Knight, M.J, Ruaux, A, Mikolajek, H, Wong-Fat-Sang, N, Withers, J, Gill, R, Wood, S.P, Wood, M, Fox, G.C, Cooper, J.B. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Structure of Bipd: An Invasion Protein Associated with the Type III Secretion System of Burkholderia Pseudomallei.
J.Mol.Biol., 363, 2006
|
|
2IUM
 
 | |
2IUN
 
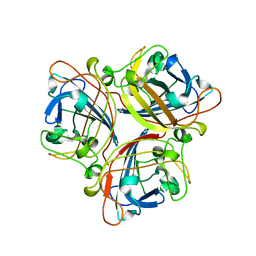 | | Structure of the C-terminal head domain of the avian adenovirus CELO long fibre (P21 crystal form) | | Descriptor: | AVIAN ADENOVIRUS CELO LONG FIBRE, CALCIUM ION | | Authors: | Guardado-Calvo, P, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2006-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the C-terminal head domain of the fowl adenovirus type 1 long fiber.
J. Gen. Virol., 88, 2007
|
|
2BT8
 
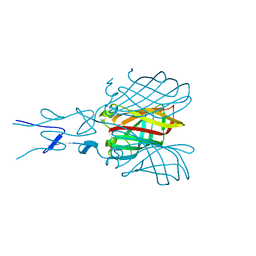 | | Structure of the C-terminal receptor-binding domain of avian reovirus fibre sigmaC, space group P6322. | | Descriptor: | SIGMA C | | Authors: | Guardado Calvo, P, Fox, G.C, Hermo Parrado, X.L, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Carboxy-Terminal Receptor-Binding Domain of Avian Reovirus Fibre Sigmac
J.Mol.Biol., 354, 2005
|
|
2R9L
 
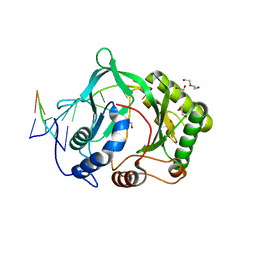 | | Polymerase Domain from Mycobacterium tuberculosis Ligase D in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*DGP*DCP*DCP*DGP*DCP*DAP*DAP*DCP*DGP*DCP*DA)-3'), ... | | Authors: | Brissett, N.C, Fox, G.C, Pitcher, R.S, Doherty, A.J. | | Deposit date: | 2007-09-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a NHEJ polymerase-mediated DNA synaptic complex
Science, 318, 2007
|
|
2W37
 
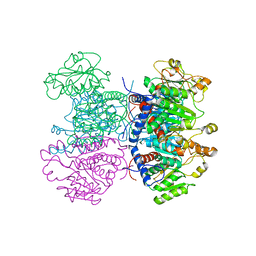 | | CRYSTAL STRUCTURE OF THE HEXAMERIC CATABOLIC ORNITHINE TRANSCARBAMYLASE FROM Lactobacillus hilgardii | | Descriptor: | NICKEL (II) ION, ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | de las Rivas, B, Fox, G.C, Angulo, I, Rodriguez, H, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hexameric Catabolic Ornithine Transcarbamylase from Lactobacillus Hilgardii: Structural Insights Into the Oligomeric Assembly and Metal Binding.
J.Mol.Biol., 393, 2009
|
|
2XB8
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase in complex with inhibitor compound (2R)-2-(4-methoxybenzyl)-3- dehydroquinic acid | | Descriptor: | (1R,2R,4S,5R)-1,4,5-TRIHYDROXY-2-(4-METHOXYBENZYL)-3-OXOCYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Tizon, L, Llamas-Saiz, A.L, Fox, G.C, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the Key Factors that Control the Inhibition of Type II Dehydroquinase by (2R)-2- Benzyl-3-Dehydroquinic Acids.
Chemmedchem, 5, 2010
|
|
2XB9
 
 | | Structure of Helicobacter pylori type II dehydroquinase in complex with inhibitor compound (2R)-2-(4-methoxybenzyl)-3-dehydroquinic acid | | Descriptor: | (1R,2R,4S,5R)-1,4,5-TRIHYDROXY-2-(4-METHOXYBENZYL)-3-OXOCYCLOHEXANECARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CITRIC ACID | | Authors: | Otero, J.M, Tizon, L, Llamas-Saiz, A.L, Fox, G.C, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Understanding the Key Factors that Control the Inhibition of Type II Dehydroquinase by (2R)-2- Benzyl-3-Dehydroquinic Acids.
Chemmedchem, 5, 2010
|
|
2XDA
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-2-(2-Cyclopropyl)ethyl-4,6,7- trihydroxy-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-2-(2-CYCLOPROPYLETHYL)-4,6,7-TRIHYDROXY-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
2XD9
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-4,6,7-Trihydroxy-2-((E)-prop-1- enyl)-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-4,6,7-TRIHYDROXY-2-[(1E)-PROP-1-EN-1-YL]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Lapthorn, A.J, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
3PKY
 
 | | Polymerase Domain from Mycobacterium tuberculosis Ligase D in complex with DNA, UTP and Manganese. | | Descriptor: | DNA 5'-D(*G*CP*CP*GP*CP*AP*AP*CP*GP*CP*AP*CP*G)-3', DNA 5'-D(P*GP*CP*GP*GP*C)-3', MANGANESE (II) ION, ... | | Authors: | Brissett, N.C, Fox, G.C, Pitcher, R.S, Doherty, A.J. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a Preternary Complex Involving a Prokaryotic NHEJ DNA Polymerase.
Mol.Cell, 41, 2011
|
|
2J16
 
 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2J17
 
 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
6Q3T
 
 | | Structure of Protease1 from Pyrococcus horikoshii at room temperature in ChipX microfluidic device | | Descriptor: | Deglycase PH1704 | | Authors: | de Wijn, R, Engilberge, S, Olieric, V, Girard, E, Sauter, C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6Q52
 
 | | Structure of a psychrophilic CCA-adding enzyme in complex with CMPcPP at room temperature in ChipX microfluidic device | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-12-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
4LIT
 
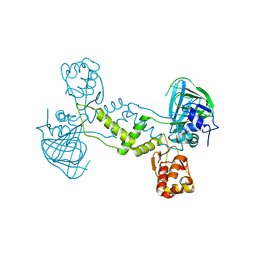 | |
4LIV
 
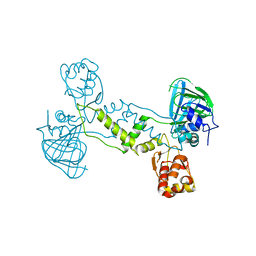 | |
5ZMI
 
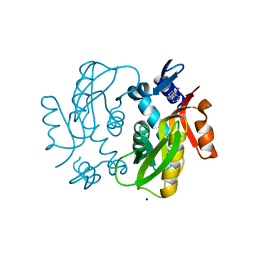 | |
6GZP
 
 | | Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic device | | Descriptor: | Nanobody | | Authors: | Roche, J, Gaubert, A, Desmyter, A, De Wijn, R, Sauter, C, Roussel, A. | | Deposit date: | 2018-07-04 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6HW1
 
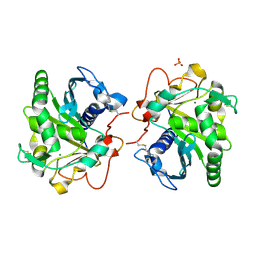 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6IBQ
 
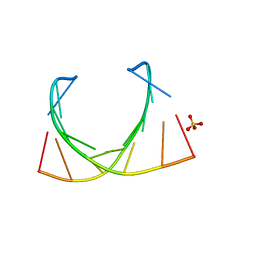 | | Structure of a nonameric RNA duplex at room temperature in ChipX microfluidic device | | Descriptor: | DNA/RNA (5'-R(*CP*GP*UP*GP*AP*UP*CP*G)-D(P*C)-3'), SULFATE ION | | Authors: | de Wijn, R, Olieric, V, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6IBP
 
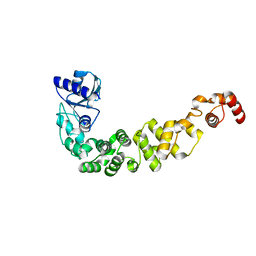 | | Structure of a psychrophilic CCA-adding enzyme at room temperature in ChipX microfluidic device | | Descriptor: | CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
2IRX
 
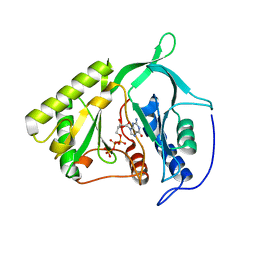 | |
