4QX2
 
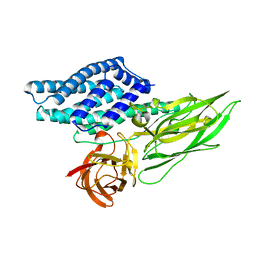 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX1
 
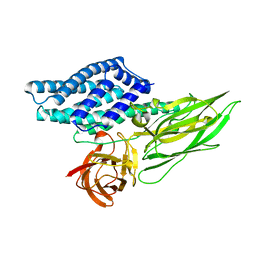 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3G5O
 
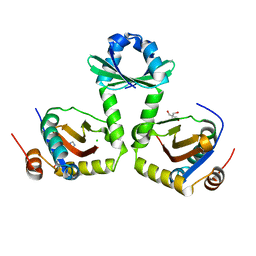 | | The crystal structure of the toxin-antitoxin complex RelBE2 (Rv2865-2866) from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-14 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
4M5S
 
 | | Human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SUCCINIC ACID | | Authors: | Laganowsky, A, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MJH
 
 | |
4QXX
 
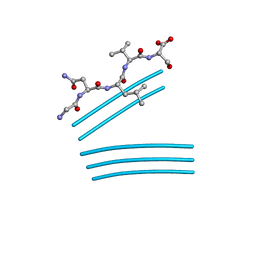 | |
4M5T
 
 | | Disulfide trapped human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Cascio, D, Hochberg, G, Sawaya, M.R, Benesch, J.L.P, Robinson, C.V, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5IIW
 
 | | Corkscrew assembly of SOD1 residues 28-38 without potassium iodide | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CYU
 
 | | Structure of the soluble domain of EccB1 from the Mycobacterium smegmatis ESX-1 secretion system. | | Descriptor: | Conserved membrane protein | | Authors: | Arbing, M.A, Chan, S, Kahng, S, Kim, J, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system.
Bmc Struct.Biol., 16, 2016
|
|
1I8F
 
 | | THE CRYSTAL STRUCTURE OF A HEPTAMERIC ARCHAEAL SM PROTEIN: IMPLICATIONS FOR THE EUKARYOTIC SNRNP CORE | | Descriptor: | GLYCEROL, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Mura, C, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2001-03-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8T84
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes hexafluoroisopropanol | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, Racemic mixture of amyloid beta segment 35-MVGGVV-40 | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T86
 
 | |
8T89
 
 | |
8T82
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes pentafluoropropionic acid | | Descriptor: | amyloid beta segment 35-MVGGVV-40, racemic mixture, pentafluoropropanoic acid | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
6PQ5
 
 | |
6PQA
 
 | |
3OEI
 
 | | Crystal structure of Mycobacterium tuberculosis RelJK (Rv3357-Rv3358-RelBE3) | | Descriptor: | CITRATE ANION, RelJ (Antitoxin Rv3357), RelK (Toxin Rv3358) | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
4RCL
 
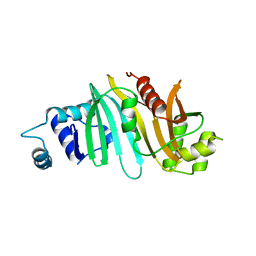 | |
5TY4
 
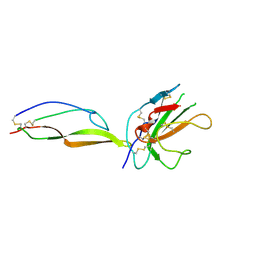 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5DLI
 
 | | Corkscrew assembly of SOD1 residues 28-38 | | Descriptor: | GLYCEROL, IODIDE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2015-09-05 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4KK7
 
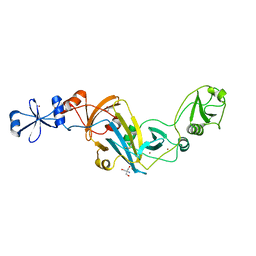 | |
4NIN
 
 | |
4ONK
 
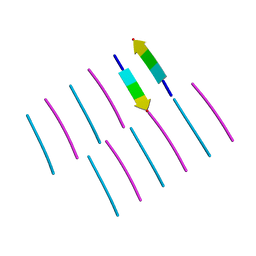 | | [Leu-5]-Enkephalin mutant - YVVFL | | Descriptor: | [Leu-5]-Enkephalin mutant - YVVFL | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
4OLR
 
 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
1JCV
 
 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE LOW TEMPERATURE (-180C) STRUCTURE | | Descriptor: | COPPER (II) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Ogihara, N.L, Parge, H.E, Hart, P.J, Weiss, M.S, Valentine, J.S, Eisenberg, D.S, Tainer, J.A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
