7UBN
 
 | |
7UBL
 
 | | Transcription antitermination factor Qlambda in complex with Q-lambda-binding-element DNA | | 分子名称: | 1,2-ETHANEDIOL, Antitermination protein Q, DNA (5'-D(P*CP*AP*CP*CP*CP*AP*AP*TP*TP*TP*TP*AP*TP*TP*CP*AP*AP*TP*G)-3'), ... | | 著者 | Yin, Z, Ebright, R.H. | | 登録日 | 2022-03-15 | | 公開日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (2.177 Å) | | 主引用文献 | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1O3R
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3Q
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3T
 
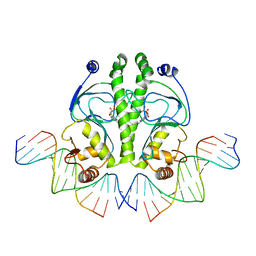 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex:
DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3S
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | 分子名称: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | 著者 | Chen, S, Ebright, R.H, Berman, H.M. | | 登録日 | 2003-03-18 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: Alteration of DNA Binding Specificity Through Alteration of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
5D4D
 
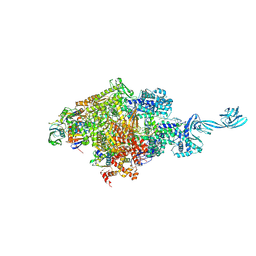 | |
5D4C
 
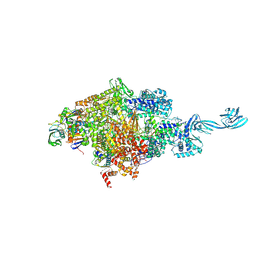 | |
5D4E
 
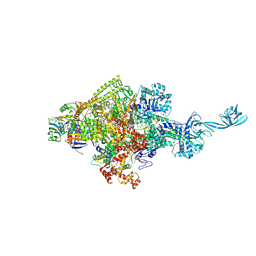 | |
5E17
 
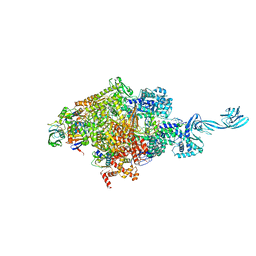 | |
5E18
 
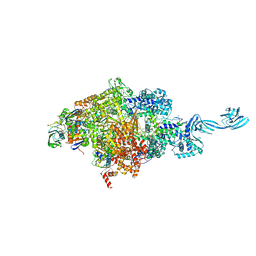 | |
1PP5
 
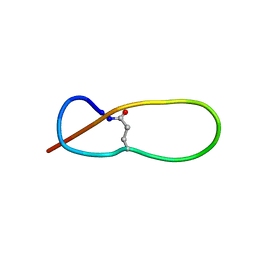 | | Structure of Antibacterial Peptide Microcin J25: a 21-Residue Lariat Protoknot | | 分子名称: | microcin J25 | | 著者 | Bayro, M.J, Swapna, G.V.T, Huang, J.Y, Ma, L.-C, Mukhopadhyay, J, Ebright, R.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-06-16 | | 公開日 | 2003-10-28 | | 最終更新日 | 2012-12-12 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Antibacterial Peptide Microcin J25: A 21-Residue Lariat Protoknot.
J.Am.Chem.Soc., 125, 2003
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | 著者 | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | 登録日 | 1996-05-26 | | 公開日 | 1997-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1RUO
 
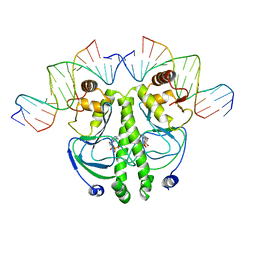 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | 著者 | Parkinson, G.N, Ebright, R.H, Berman, H.M. | | 登録日 | 1996-05-26 | | 公開日 | 1997-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X7K
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XGF
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-17 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X9Q
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 27 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-03 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-20 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X6T
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B1 (TTC-B1) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XIJ
 
 | | Escherichia coli transcription-translation complex A (TTC-A) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-20 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDR
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 27 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8E70
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, dC75 rut mimic RNA, Mg-ADP-BeF3, and NusG; Rho hexamer part | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Molodtsov, V, Wang, C, Ebright, R.H. | | 登録日 | 2022-08-23 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E6Z
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, dC75 rut mimic RNA, Mg-ADP-BeF3, and NusG; TEC part | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Molodtsov, V, Wang, C, Ebright, R.H. | | 登録日 | 2022-08-23 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
