4V2D
 
 | | FLRT2 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4UU9
 
 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | Descriptor: | COMPLEMENT C5, MEDI7814, SULFATE ION | | Authors: | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4V8A
 
 | | The structure of thermorubin in complex with the 70S ribosome from Thermus thermophilus. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D, Johnson, F.A, Steitz, T.A. | | Deposit date: | 2011-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge b2a of the ribosome.
J.Mol.Biol., 416, 2012
|
|
4V0V
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-660) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4URJ
 
 | | Crystal structure of human BJ-TSA-9 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN FAM83A | | Authors: | Pinkas, D.M, Sanvitale, C, Wang, D, Krojer, T, Kopec, J, Chaikuad, A, Dixon Clarke, S, Berridge, G, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of Human Bj-Tsa-9
To be Published
|
|
4UZM
 
 | |
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
3CLV
 
 | | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Rab5 protein, ... | | Authors: | Chattopadhyay, D, Wernimont, A.K, Langsley, G, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c
To be Published
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
1N4H
 
 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | Nuclear Receptor ROR-beta, RETINOIC ACID, Steroid Receptor Coactivator-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2002-10-31 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
6XNS
 
 | | C3_crown-05 | | Descriptor: | C3_crown-05 | | Authors: | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XT4
 
 | |
6X79
 
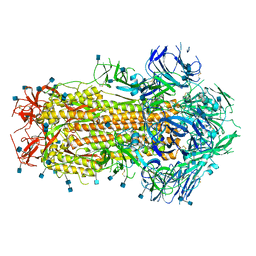 | | Prefusion SARS-CoV-2 S ectodomain trimer covalently stabilized in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | McCallum, M, Walls, A.C, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-19 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1EPH
 
 | |
1N15
 
 | |
1N50
 
 | |
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILV
 
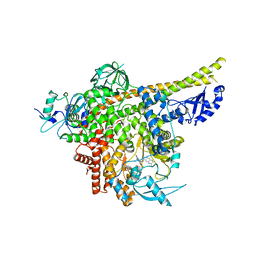 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILS
 
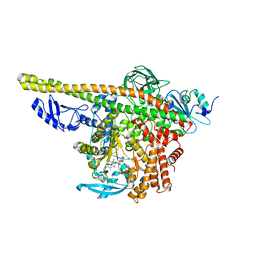 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3CPO
 
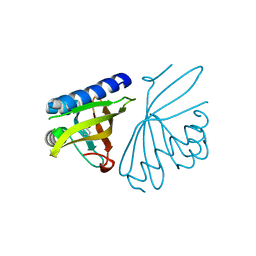 | | Crystal structure of ketosteroid isomerase D40N with bound 2-fluorophenol | | Descriptor: | 2-fluorophenol, Delta(5)-3-ketosteroid isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole
J.Am.Chem.Soc., 130, 2008
|
|
1EVV
 
 | |
1EEH
 
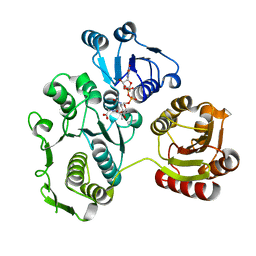 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
6XYE
 
 | |
