2CCM
 
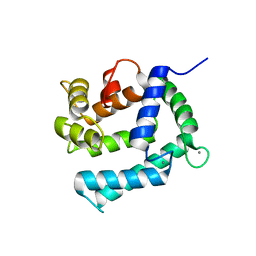 | | X-ray structure of Calexcitin from Loligo pealeii at 1.8A | | Descriptor: | CALCIUM ION, CALEXCITIN | | Authors: | Erskine, P.T, Beaven, G.D.E, Wood, S.P, Fox, G, Vernon, J, Giese, K.P, Cooper, J.B. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Neuronal Protein Calexcitin Suggests a Mode of Interaction in Signalling Pathways of Learning and Memory.
J.Mol.Biol., 357, 2006
|
|
5K4U
 
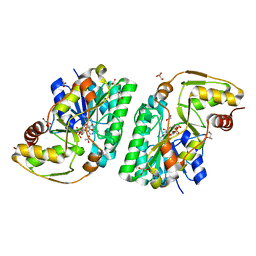 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei showing different active site loop conformations between dimer subunits, refined to 1.9 angstroms | | Descriptor: | ACETATE ION, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.K, Cooper, J.B, Erskine, P.T. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5K4Q
 
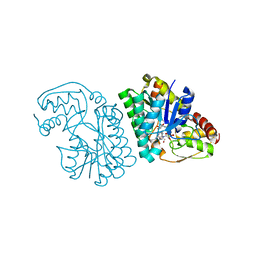 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ refined to 2.3 angstroms | | Descriptor: | GLYCEROL, L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Adjogatse, E.K, Cooper, J.B, Erskine, P.T. | | Deposit date: | 2016-05-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5K4T
 
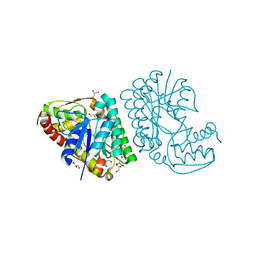 | |
5K4V
 
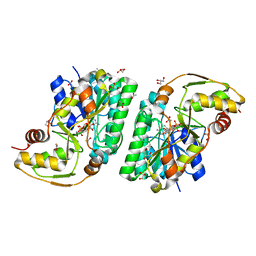 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ refined to 2.2 angstroms | | Descriptor: | ACETATE ION, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5K50
 
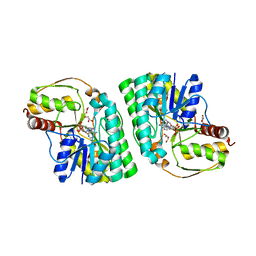 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ and L-allo-threonine refined to 2.23 angstroms | | Descriptor: | ACETATE ION, ALLO-THREONINE, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5K4Y
 
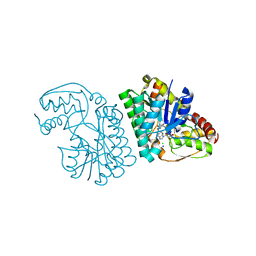 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei refined to 1.77 angstroms | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5KMW
 
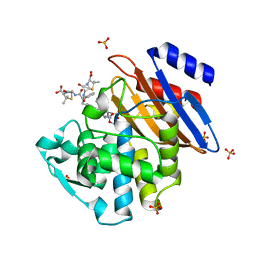 | | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | | Descriptor: | Beta-lactamase Toho-1, OPEN FORM - PENICILLIN G, PENICILLIN G, ... | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Ginell, S.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex
to be published
|
|
5KSC
 
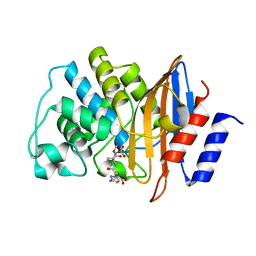 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | Authors: | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-04 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
5L9A
 
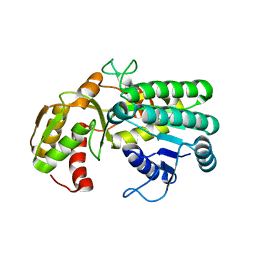 | | L-threonine dehydrogenase from trypanosoma brucei. | | Descriptor: | ACETATE ION, L-threonine 3-dehydrogenase | | Authors: | Erskine, P.T, Cooper, J.B, Adjogatse, E, Kelly, J, Wood, S.P. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5LZL
 
 | | Pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase | | Descriptor: | Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Azim, N, Erskine, P.T, Guo, J, Cooper, J.B. | | Deposit date: | 2016-09-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LC1
 
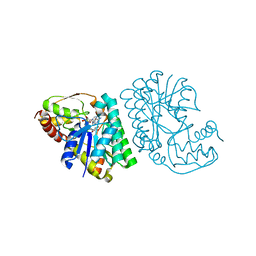 | | L-threonine dehydrogenase from Trypanosoma brucei with NAD and the inhibitor pyruvate bound. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, L-threonine 3-dehydrogenase, ... | | Authors: | Erskine, P.T, Adjogatse, E, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5MDN
 
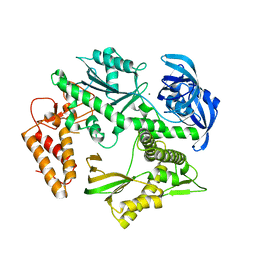 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1QNV
 
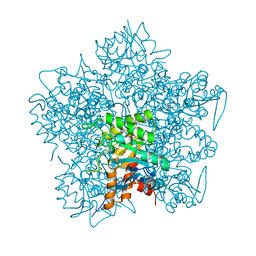 | | yeast 5-aminolaevulinic acid dehydratase Lead (Pb) complex | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LEAD (II) ION | | Authors: | Erskine, P.T, Senior, N.M, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QML
 
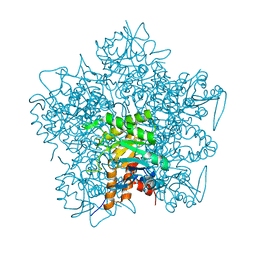 | | Hg complex of yeast 5-aminolaevulinic acid dehydratase | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, MERCURY (II) ION | | Authors: | Erskine, P.T, Senior, N, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-02 | | Release date: | 2000-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QN2
 
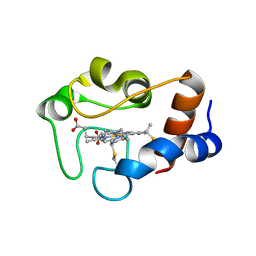 | | cytochrome cH from Methylobacterium extorquens | | Descriptor: | CYTOCHROME CH, HEME C | | Authors: | Read, J, Gill, R, Dales, S.L, Cooper, J.B, Wood, S.P, Anthony, C. | | Deposit date: | 1999-10-13 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Molecular Structure of an Unusual Cytochrome C2 Determined at 2.0A; the Cytochrome cH from Methylobacterium Extorquens
Protein Sci., 8, 1999
|
|
1EPO
 
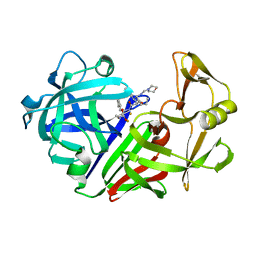 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
1EPQ
 
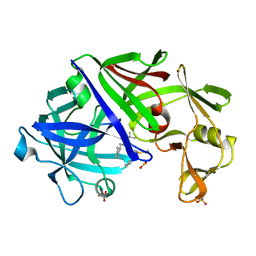 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-133,450 (SOT PHE GLY+SCC GCL) | | Descriptor: | ENDOTHIAPEPSIN, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morpholin-4-ylsulfonyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Dealwis, C, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EPP
 
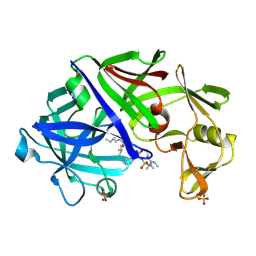 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
4HTG
 
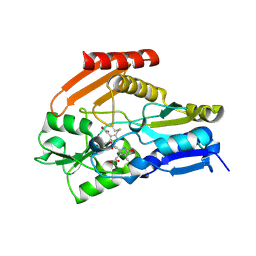 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7OL1
 
 | |
1OEX
 
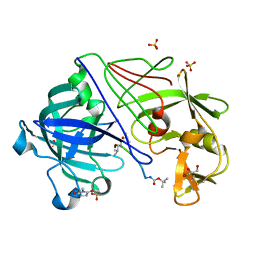 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEW
 
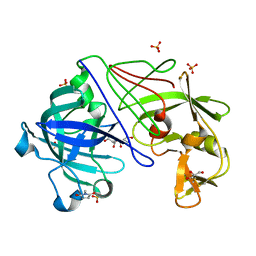 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OD1
 
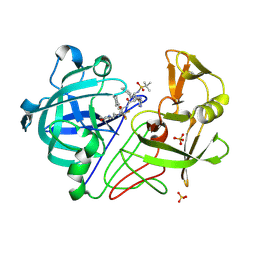 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2012-11-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OHL
 
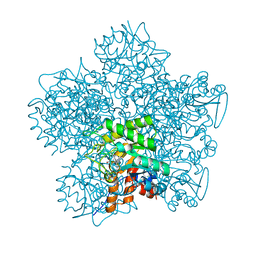 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE PUTATIVE CYCLIC REACTION INTERMEDIATE COMPLEX | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, BETA-MERCAPTOETHANOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ... | | Authors: | Erskine, P.T, Coates, L, Butler, D, Youell, J.H, Brindley, A.A, Wood, S.P, Warren, M.J, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Structure of a Putative Reaction Intermediateof 5-Aminolaevulinic Acid Dehydratase
Biochem.J., 373, 2003
|
|
