7OS4
 
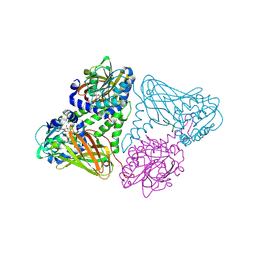 | | Crystal structure of mouse CARM1 in complex with histone H3_13-31 K18 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.1, Histone-arginine methyltransferase CARM1 | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
7OKP
 
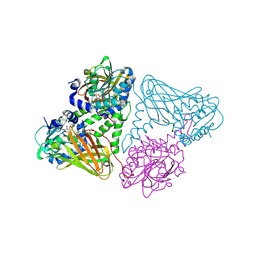 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
2XPP
 
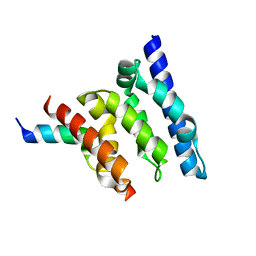 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form III | | Descriptor: | CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XPO
 
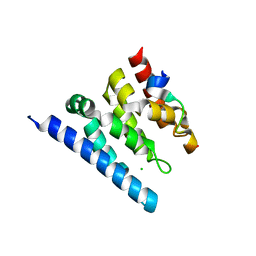 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form II | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Moras, D, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XPN
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form I | | Descriptor: | BROMIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2J49
 
 | | Crystal structure of yeast TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 5 | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
2J4B
 
 | | Crystal structure of Encephalitozoon cuniculi TAF5 N-terminal domain | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID SUBUNIT 72/90-100 KDA | | Authors: | Romier, C, James, N, Birck, C, Cavarelli, J, Vivares, C, Collart, M.A, Moras, D. | | Deposit date: | 2006-08-28 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure, Biochemical and Genetic Characterization of Yeast and E. Cuniculi Taf(II)5 N-Terminal Domain: Implications for TFIID Assembly.
J.Mol.Biol., 368, 2007
|
|
7PUC
 
 | | CARM1 in complex with EML981 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[(~{E})-3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]prop-2-enyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PU8
 
 | | CARM1 in complex with EML980 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[N-[2-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PV6
 
 | | CARM1 in complex with EML734 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, TETRAETHYLENE GLYCOL, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PUQ
 
 | | CARM1 in complex with EML982 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]propyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7QRD
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
7QPH
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_22-31 K27 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone H3 22-31 K27 acetylated, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-04 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
7P2R
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML980 | | Descriptor: | Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, methyl 6-[4-[[N-[2-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-07-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
5TBJ
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1452 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5TBH
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1236 | | Descriptor: | (2~{R})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]amino]-2-azanyl-butanoic acid, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5TBI
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1427 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-[2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]ethylamino]-1-(methoxymethyl)pyrimidin-2-one, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
2XP1
 
 | | Structure of the tandem SH2 domains from Antonospora locustae transcription elongation factor Spt6 | | Descriptor: | CHLORIDE ION, SPT6, SULFATE ION | | Authors: | Diebold, M.-L, Koch, M, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Noncanonical Tandem Sh2 Enables Interaction of Elongation Factor Spt6 with RNA Polymerase II.
J.Biol.Chem., 285, 2010
|
|
1PGL
 
 | | BEAN POD MOTTLE VIRUS (BPMV), MIDDLE COMPONENT | | Descriptor: | 5'-R(*AP*GP*UP*CP*UP*C)-3', BEAN POD MOTTLE VIRUS LARGE (L) SUBUNIT, BEAN POD MOTTLE VIRUS SMALL (S) SUBUNIT | | Authors: | Lin, T, Cavarelli, J, Johnson, J.E. | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus.
Virology, 314, 2003
|
|
1PGW
 
 | | BEAN POD MOTTLE VIRUS (BPMV), TOP COMPONENT | | Descriptor: | BEAN POD MOTTLE VIRUS LARGE (L) SUBUNIT, BEAN POD MOTTLE VIRUS SMALL (S) SUBUNIT | | Authors: | Lin, T, Cavarelli, J, Johnson, J.E. | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus.
Virology, 314, 2003
|
|
6TQS
 
 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the conventional FFAT motif of ORP1. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQU
 
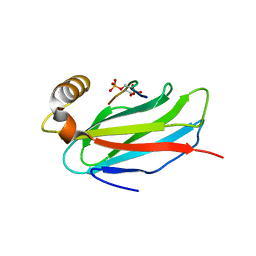 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | Motile sperm domain-containing protein 2, SULFATE ION, StAR-related lipid transfer protein 3 | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQR
 
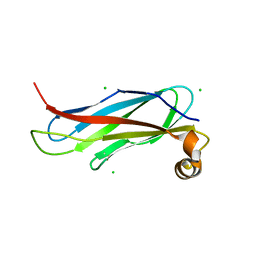 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6TQT
 
 | | The crystal structure of the MSP domain of human MOSPD2. | | Descriptor: | 1,2-ETHANEDIOL, Motile sperm domain-containing protein 2, PHOSPHATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
5G02
 
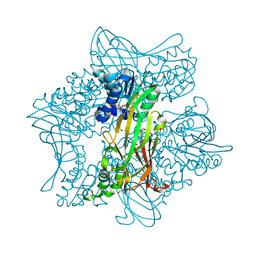 | | Crystal Structure of zebrafish Protein Arginine Methyltransferase 2 with SFG | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LITHIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-16 | | Release date: | 2016-11-09 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
