3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
1RKC
 
 | | Human vinculin head (1-258) in complex with talin's vinculin binding site 3 (residues 1944-1969) | | Descriptor: | Talin, Vinculin | | Authors: | Izard, T, Evans, G, Borgon, R.A, Rush, C.L, Bricogne, G, Bois, P.R. | | Deposit date: | 2003-11-21 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Vinculin activation by talin through helical bundle conversion
Nature, 427, 2004
|
|
1RKE
 
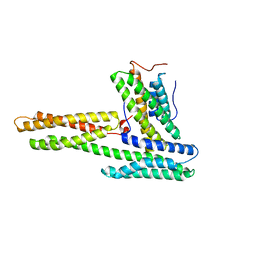 | | Human vinculin head (1-258) in complex with human vinculin tail (879-1066) | | Descriptor: | VCL protein, Vinculin | | Authors: | Izard, T, Evans, G, Borgon, R.A, Rush, C.L, Bricogne, G, Bois, P.R. | | Deposit date: | 2003-11-21 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Vinculin activation by talin through helical bundle conversion
Nature, 427, 2004
|
|
1GW9
 
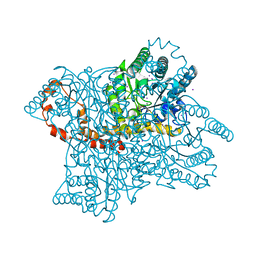 | |
1GWA
 
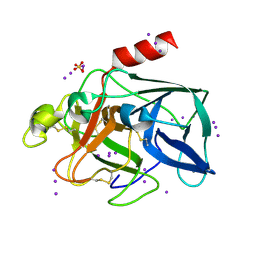 | | Triiodide derivative of porcine pancreas elastase | | Descriptor: | CALCIUM ION, ELASTASE 1, IODIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-13 | | Release date: | 2002-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GWD
 
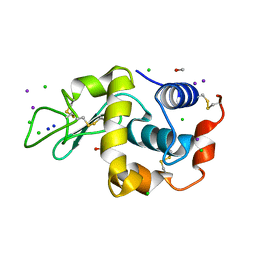 | | Tri-iodide derivative of hen egg-white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GWG
 
 | | Tri-iodide derivative of apoferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN, IODIDE ION | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-15 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2W2G
 
 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2WCT
 
 | | human SARS coronavirus unique domain (triclinic form) | | Descriptor: | NON-STRUCTURAL PROTEIN 3 | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
1TR2
 
 | | Crystal structure of human full-length vinculin (residues 1-1066) | | Descriptor: | VINCULIN ISOFORM 1 | | Authors: | Borgon, R.A, Vonrhein, C, Bricogne, G, Bois, P.R, Izard, T. | | Deposit date: | 2004-06-19 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Vinculin
Structure, 12, 2004
|
|
1UOY
 
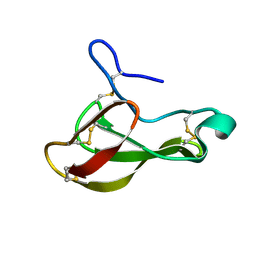 | | The bubble protein from Penicillium brevicompactum Dierckx exudate. | | Descriptor: | BUBBLE PROTEIN | | Authors: | Olsen, J.G, Flensburg, C, Olsen, O, Seibold, M, Bricogne, G, Henriksen, A. | | Deposit date: | 2003-09-26 | | Release date: | 2003-11-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Solving the Structure of the Bubble Protein Using the Anomalous Sulfur Signal from Single-Crystal in-House Cu Kalpha Diffraction Data Only
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5D98
 
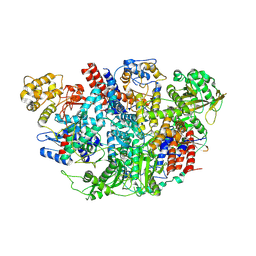 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
5D9A
 
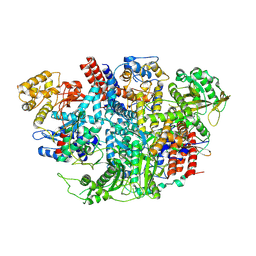 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P212121 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
1HXH
 
 | | COMAMONAS TESTOSTERONI 3BETA/17BETA HYDROXYSTEROID DEHYDROGENASE | | Descriptor: | 3BETA/17BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Benach, J, Filling, C, Oppermann, U.C.T, Roversi, P, Bricogne, G, Berndt, K.D, Jornvall, H, Ladenstein, R. | | Deposit date: | 2001-01-15 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of Bacterial 3beta/17beta-Hydroxysteroid Dehydrogenase at 1.2 A Resolution: A Model for
Multiple Steroid Recognition
Biochemistry, 41, 2002
|
|
4Z9L
 
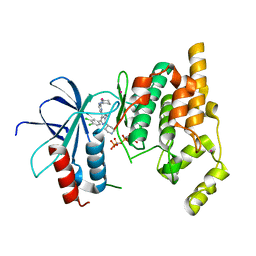 | | THE STRUCTURE OF JNK3 IN COMPLEX WITH AN IMIDAZOLE-PYRIMIDINE INHIBITOR | | Descriptor: | CYCLOHEXYL-{4-[5-(3,4-DICHLOROPHENYL)-2-PIPERIDIN-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, Lograsso, P.V, Smart, O.S, Bricogne, G. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity.
Chem.Biol., 10, 2003
|
|
1BX7
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
6CDS
 
 | | Human neurofibromin 2/merlin/schwannomin residues 1-339 in complex with PIP2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Merlin, ... | | Authors: | Chinthalapudi, K, Sharff, A.J, Bricogne, G, Izard, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2.
Nat Commun, 9, 2018
|
|
1BX8
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1LF1
 
 | | Crystal Structure of Cel5 from Alkalophilic Bacillus sp. | | Descriptor: | Cel5 | | Authors: | Shaw, A, Bott, R, Vonrhein, C, Bricogne, G, Power, S, Day, A.G. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel combination of two classic catalytic schemes.
J.Mol.Biol., 320, 2002
|
|
1MAW
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with ATP in an Open Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1LZ8
 
 | | LYSOZYME PHASED ON ANOMALOUS SIGNAL OF SULFURS AND CHLORINES | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Dauter, Z, Dauter, M, De La Fortelle, E, Bricogne, G, Sheldrick, G.M. | | Deposit date: | 1999-03-14 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Can anomalous signal of sulfur become a tool for solving protein crystal structures?
J.Mol.Biol., 289, 1999
|
|
1M83
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with ATP in a Closed, Pre-transition State Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-07-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MAU
 
 | | Crystal structure of Tryptophanyl-tRNA Synthetase Complexed with ATP and Tryptophanamide in a Pre-Transition state Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, GLYCEROL, ... | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MB2
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with Tryptophan in an Open Conformation | | Descriptor: | TRYPTOPHAN, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
