4J8F
 
 | |
4J8D
 
 | |
3ZUH
 
 | | Negative stain EM Map of the AAA protein CbbX, a red-type Rubisco activase from R. sphaeroides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN CBBX, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-09 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Structure and Function of the Aaa+ Protein Cbbx, a Red-Type Rubisco Activase.
Nature, 479, 2011
|
|
3ZW6
 
 | | MODEL OF HEXAMERIC AAA DOMAIN ARRANGEMENT OF GREEN-TYPE RUBISCO ACTIVASE FROM TOBACCO. | | Descriptor: | RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE ACTIVASE 1, CHLOROPLASTIC | | Authors: | Stotz, M, Mueller-Cajar, O, Ciniawsky, S, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-09 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of Green-Type Rubisco Activase from Tobacco
Nat.Struct.Mol.Biol., 18, 2011
|
|
5D5W
 
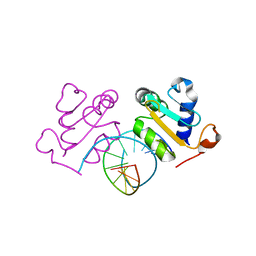 | | Crystal structure of Chaetomium thermophilum Skn7 with HSE DNA | | Descriptor: | HSE DNA, Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Z
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form II | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Bracher, A, Hartl, F.U. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Y
 
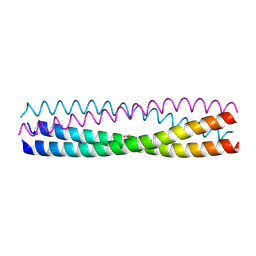 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5DIT
 
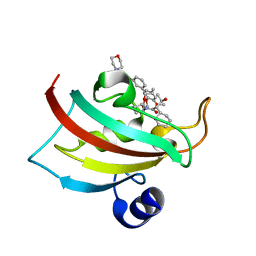 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2015-10-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
5D5V
 
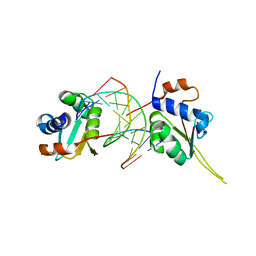 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5X
 
 | | Crystal structure of Chaetomium thermophilum Skn7 with SSRE DNA | | Descriptor: | Putative transcription factor, SSRE DNA strand 1, SSRE DNA strand 2 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5U
 
 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D60
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form III | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4DRO
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH (1R)-3-(3,4-dimethoxyphenyl)-1-phenylpropyl (2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRH
 
 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR at low pH | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DRK
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRM
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRJ
 
 | | o-crystal structure of the PPIase domain of FKBP52, Rapamycin and the FRB fragment of mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRN
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRI
 
 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
4DRP
 
 | | Evaluation of Synthetic FK506 Analogs as Ligands for the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-3-(3,4-dimethoxyphenyl)-1-((S)-1-(2-((1R,2S)-2-ethyl-1-hydroxy-cyclohexyl)-2-oxoacetyl)piperidine-2-carbonyloxy)propyl)phenoxy)acetic acid from cocrystallization | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
6Z1E
 
 | |
6Z1D
 
 | |
6Z1F
 
 | | CryoEM structure of Rubisco Activase with its substrate Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
6YUN
 
 | |
