4CGB
 
 | | Crystal structure of the trimerization domain of EML2 | | Descriptor: | ECHINODERM MICROTUBULE-ASSOCIATED PROTEIN-LIKE 2, GLYCEROL, POTASSIUM ION | | Authors: | Richards, M.W, Bayliss, R. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Microtubule Association of Eml Proteins and the Eml4-Alk Variant 3 Oncoprotein Require an N-Terminal Trimerization Domain.
Biochem.J., 467, 2015
|
|
4CEG
 
 | | Crystal structure of Aurora A 122-403 C290A, C393A bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, CHLORIDE ION, ... | | Authors: | Burgess, S.G, Bayliss, R. | | Deposit date: | 2013-11-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of C290A:C393A Aurora a Provides Structural Insights Into Kinase Regulation.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4B0G
 
 | | Complex of Aurora-A bound to an Imidazopyridine-based inhibitor | | Descriptor: | 6-bromo-2-(1-methyl-1H-imidazol-5-yl)-7-{4-[(5-methyl-1,2-oxazol-3-yl)methyl]piperazin-1-yl}-1H-imidazo[4,5-b]pyridine, AURORA KINASE A, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2012-07-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of Imidazo[4,5-B]Pyridine-Based Kinase Inhibitors: Identification of a Dual Flt3/Aurora Kinase Inhibitor as an Orally Bioavailable Preclinical Development Candidate for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 55, 2012
|
|
4CVA
 
 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
4CV8
 
 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 3-amino-5-(1-methyl-1H-pyrazol-4-yl)pyridin-2(1H)-one, DUAL SPECIFICITY PROTEIN KINASE TTK | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
4CI8
 
 | |
4CGC
 
 | |
2XK4
 
 | | Structure of Nek2 bound to aminopyrazine compound 17 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-ethoxybenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNO
 
 | | Structure of Nek2 bound to CCT243779 | | Descriptor: | 1,2-ETHANEDIOL, 5-{6-[(1-methylpiperidin-4-yl)oxy]-1H-benzimidazol-1-yl}-3-{[2-(trifluoromethyl)benzyl]oxy}thiophene-2-carboxamide, CHLORIDE ION, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
2XNP
 
 | | Structure of Nek2 bound to CCT244858 | | Descriptor: | 1,2-ETHANEDIOL, 4-{5-[(1-METHYLPIPERIDIN-4-YL)OXY]-1H-BENZIMIDAZOL-1-YL}-2-{(1R)-1-[2-(TRIFLUOROMETHYL)PHENYL]ETHOXY}BENZAMIDE, CHLORIDE ION, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
2XK7
 
 | | Structure of Nek2 bound to aminopyrazine compound 23 | | Descriptor: | (3R,4R)-1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-3-ethylpiperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNG
 
 | | Structure of Aurora-A bound to a selective imidazopyrazine inhibitor | | Descriptor: | N-(3-{3-chloro-8-[(4-morpholin-4-ylphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzyl)methanesulfonamide, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2XKD
 
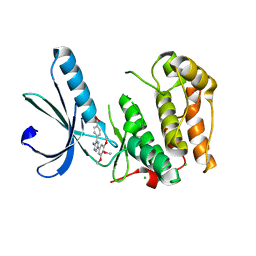 | | Structure of Nek2 bound to aminopyrazine compound 12 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]benzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKC
 
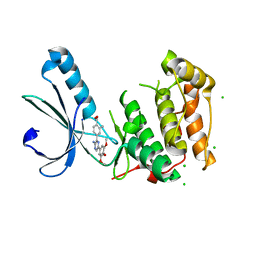 | | Structure of Nek2 bound to aminopyrazine compound 14 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-methylbenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XP0
 
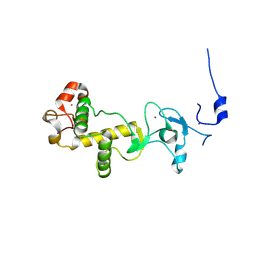 | | C-terminal cysteine-rich domain of human CHFR | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE CHFR, ZINC ION | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
2XKE
 
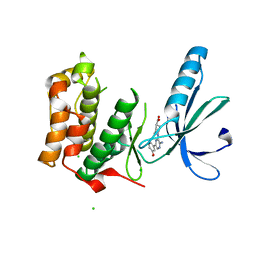 | | Structure of Nek2 bound to Aminipyrazine Compound 5 | | Descriptor: | 1-[3-amino-6-(3-methoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKF
 
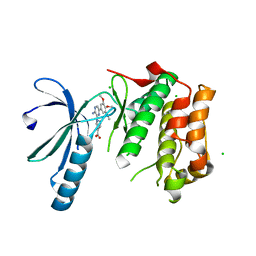 | | Structure of Nek2 bound to aminopyrazine compound 2 | | Descriptor: | 1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNN
 
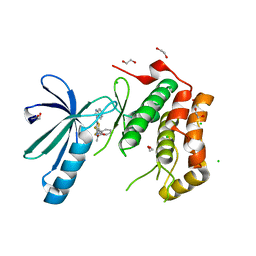 | | Structure of Nek2 bound to CCT242430 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1H-benzimidazol-1-yl)-3-{(1R)-1-[2-(trifluoromethyl)phenyl]ethoxy}thiophene-2-carboxamide, CHLORIDE ION, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
2XK8
 
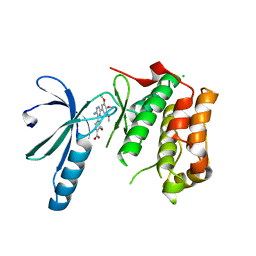 | | Structure of Nek2 bound to aminopyrazine compound 15 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-methoxybenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XOY
 
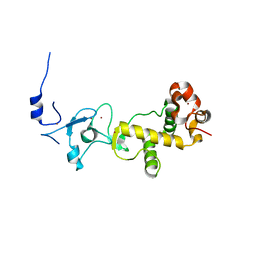 | |
2XK6
 
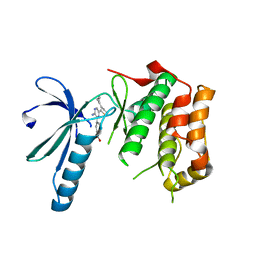 | | Structure of Nek2 bound to aminopyrazine compound 36 | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2, cis-4-[3-amino-6-(3-cyclopropylthiophen-2-yl)pyrazin-2-yl]cyclohexanecarboxylic acid | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNE
 
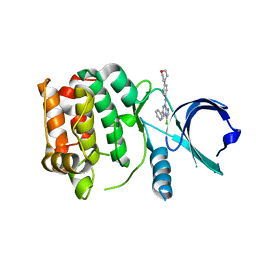 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2XOZ
 
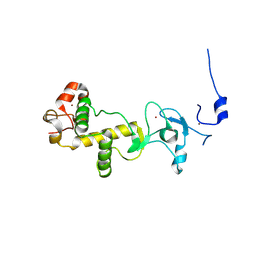 | | C-terminal cysteine rich domain of human CHFR bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ZINC ION | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
2XNM
 
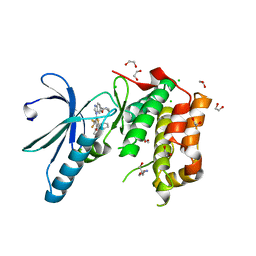 | | Structure of NEK2 bound to CCT | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-{6-[(1-METHYLPIPERIDIN-4-YL)OXY]-1H-BENZIMIDAZOL-1-YL}-3-{(1R)-1-[2-(TRIFLUOROMETHYL)PHENYL]ETHOXY}THIOPHENE-2-CARBOXAMIDE, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
2XOC
 
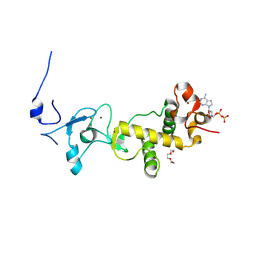 | | C-terminal cysteine-rich domain of human CHFR bound to mADPr | | Descriptor: | ADENINE, ADENOSINE-5'-DIPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ... | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
