1BC8
 
 | | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), PROTEIN (SAP-1 ETS DOMAIN), ... | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1998-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
1BGC
 
 | |
1BGE
 
 | |
1BHP
 
 | | STRUCTURE OF BETA-PUROTHIONIN AT ROOM TEMPERATURE AND 1.7 ANGSTROMS RESOLUTION | | Descriptor: | ACETATE ION, BETA-PUROTHIONIN, GLYCEROL, ... | | Authors: | Teeter, M.M, Stec, B, Rao, U. | | Deposit date: | 1995-03-15 | | Release date: | 1996-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refinement of purothionins reveals solute particles important for lattice formation and toxicity. Part 2: structure of beta-purothionin at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1BD3
 
 | | STRUCTURE OF THE APO URACIL PHOSPHORIBOSYLTRANSFERASE, 2 MUTANT C128V | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1BF0
 
 | | CALCICLUDINE (CAC) FROM GREEN MAMBA DENDROASPIS ANGUSTICEPS, NMR, 15 STRUCTURES | | Descriptor: | CALCICLUDINE | | Authors: | Gilquin, B, Lecoq, A, Desne, F, Guenneugues, M, Zinn-Justin, S, Menez, A. | | Deposit date: | 1998-05-26 | | Release date: | 1999-01-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Conformational and functional variability supported by the BPTI fold: solution structure of the Ca2+ channel blocker calcicludine.
Proteins, 34, 1999
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1BFJ
 
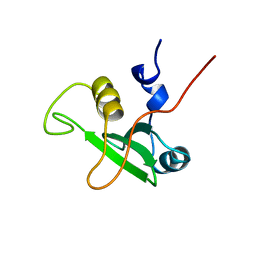 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1AX7
 
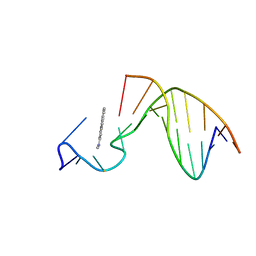 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(AAC-[AF]G-CTACCATCC)D(GGATGGTAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction.
Biochemistry, 36, 1997
|
|
1BGK
 
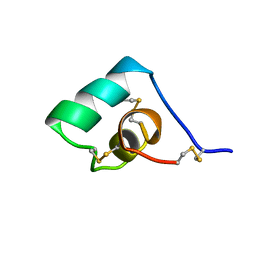 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
1B4I
 
 | | Control of K+ Channel Gating by protein phosphorylation: structural switches of the inactivation gate, NMR, 22 structures | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1AXD
 
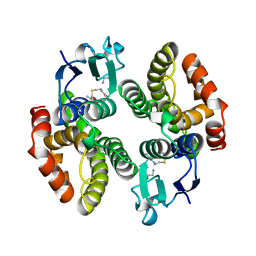 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE-I BOUND WITH THE LIGAND LACTOYLGLUTATHIONE | | Descriptor: | GLUTATHIONE S-TRANSFERASE I, LACTOYLGLUTATHIONE | | Authors: | Neuefeind, T, Huber, R, Dasenbrock, H, Prade, L, Bieseler, B. | | Deposit date: | 1997-10-15 | | Release date: | 1998-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of herbicide-detoxifying maize glutathione S-transferase-I in complex with lactoylglutathione: evidence for an induced-fit mechanism.
J.Mol.Biol., 274, 1997
|
|
1BD4
 
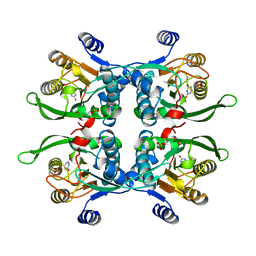 | | UPRT-URACIL COMPLEX | | Descriptor: | PHOSPHATE ION, URACIL, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1AXN
 
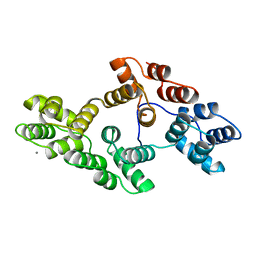 | |
1AR5
 
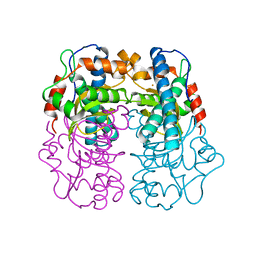 | |
1BIW
 
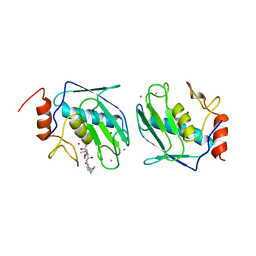 | | DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS | | Descriptor: | CALCIUM ION, N1-HYDROXY-2-(3-HYDROXY-PROPYL)-3-ISOBUTYL-N4-[1-(2-METHOXY-ETHYL)-2-OXO-AZEPAN-3-YL]-SUCCINAMIDE, PROTEIN (STROMELYSIN-1 COMPLEX), ... | | Authors: | Natchus, M.G, Cheng, M, Wahl, C.T, Pikul, S, Almstead, N.G, Bradley, R.S, Taiwo, Y.O, Mieling, G.E, Dunaway, C.M, Snider, C.E, McIver, J.M, Barnett, B.L, McPhail, S.J, Anastasio, M.B, De, B. | | Deposit date: | 1998-06-19 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of conformationally-constrained MMP inhibitors.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
1BRT
 
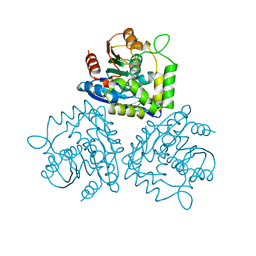 | | BROMOPEROXIDASE A2 MUTANT M99T | | Descriptor: | BROMOPEROXIDASE A2, CHLORIDE ION | | Authors: | Hofmann, B, Toelzer, S, Pelletier, I, Altenbuchner, J, Van Pee, K.H, Hecht, H.J. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation of the cofactor-free chloroperoxidases.
J.Mol.Biol., 279, 1998
|
|
1BT1
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE NATIVE CU(II)-CU(II) STATE | | Descriptor: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BUI
 
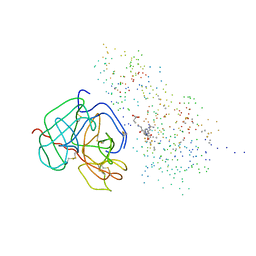 | | Structure of the ternary microplasmin-staphylokinase-microplasmin complex: a proteinase-cofactor-substrate complex in action | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, Plasminogen, Staphylokinase | | Authors: | Parry, M.A.A, Fernandez-Catalan, C, Bergner, A, Huber, R, Hopfner, K, Schlott, B, Guehrs, K, Bode, W. | | Deposit date: | 1998-09-04 | | Release date: | 1999-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The ternary microplasmin-staphylokinase-microplasmin complex is a proteinase-cofactor-substrate complex in action.
Nat.Struct.Biol., 5, 1998
|
|
1BY8
 
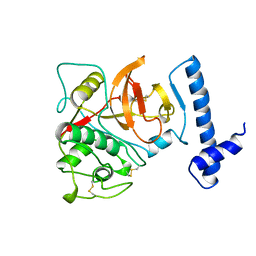 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
1BY3
 
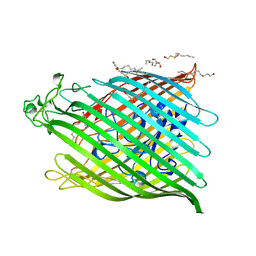 | | FHUA FROM E. COLI | | Descriptor: | N-OCTYL-2-HYDROXYETHYL SULFOXIDE, PROTEIN (FERRICHROME-IRON RECEPTOR PRECURSOR (FHUA)) | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1C3X
 
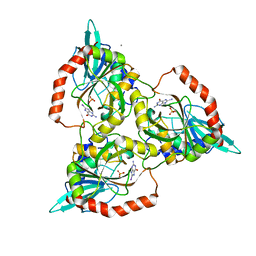 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH 8-IODO-GUANINE | | Descriptor: | 8-IODO-GUANINE, CALCIUM ION, PENTOSYLTRANSFERASE, ... | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-29 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
1BLJ
 
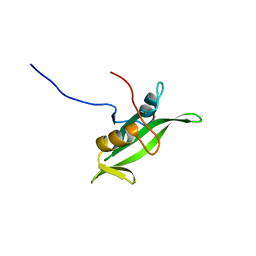 | | NMR ENSEMBLE OF BLK SH2 DOMAIN, 20 STRUCTURES | | Descriptor: | P55 BLK PROTEIN TYROSINE KINASE | | Authors: | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | Deposit date: | 1996-03-26 | | Release date: | 1997-03-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
1BT3
 
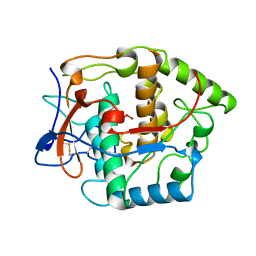 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE NATIVE CU(II)-CU(II) STATE | | Descriptor: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
