7NP9
 
 | |
4X7T
 
 | |
4X7S
 
 | |
5HLO
 
 | | Crystal structure of calcium and zinc-bound human S100A8 in space group C2221 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
4XW2
 
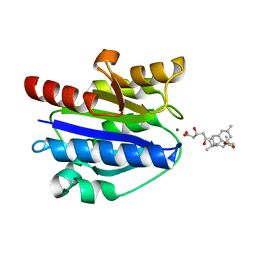 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | Descriptor: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | Authors: | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2015-01-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
6YO6
 
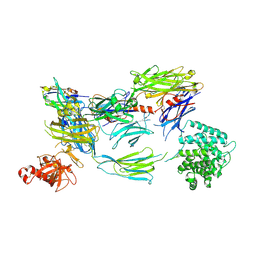 | | Structure of iC3b1 | | Descriptor: | hC3Nb1, iC3b1 alpha chain, iC3b1 beta chain | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2020-04-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Complement Receptor 3 Forms a Compact High-Affinity Complex with iC3b.
J Immunol., 206, 2021
|
|
5HLV
 
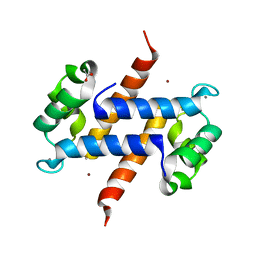 | | Crystal structure of calcium and zinc-bound human S100A8 in space group P212121 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
4LP4
 
 | |
4LP5
 
 | |
4M76
 
 | | Integrin I domain of complement receptor 3 in complex with C3d | | Descriptor: | Complement C3, Integrin alpha-M, NICKEL (II) ION | | Authors: | Bajic, G, Yatime, L, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6YSQ
 
 | | The hC4Nb8 complement inhibitory nanobody in complex with C4b | | Descriptor: | Complement C4 beta chain, Complement C4 gamma chain, Complement C4-A alpha chain, ... | | Authors: | Zarantonello, A, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | An Ultrahigh-Affinity Complement C4b-Specific Nanobody Inhibits In Vivo Assembly of the Classical Pathway Proconvertase.
J Immunol., 205, 2020
|
|
6Z6V
 
 | | Globular head of C1q in complex with the nanobody C1qNb75 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, ... | | Authors: | Laursen, N.S, Andersen, G.R. | | Deposit date: | 2020-05-29 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Functional and Structural Characterization of a Potent C1q Inhibitor Targeting the Classical Pathway of the Complement System.
Front Immunol, 11, 2020
|
|
5I5K
 
 | |
4YBH
 
 | |
5I8Q
 
 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | Authors: | He, Y, Nielsen, K.H, Andersen, G.R. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
5NQW
 
 | | IgE-Fc in complex with single domain antibody 026 | | Descriptor: | 026, GLYCEROL, Immunoglobulin heavy constant epsilon, ... | | Authors: | Laursen, N.S, Jabs, F, Spillner, E, Andersen, G.R. | | Deposit date: | 2017-04-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Trapping IgE in a closed conformation by mimicking CD23 binding prevents and disrupts Fc epsilon RI interaction.
Nat Commun, 9, 2018
|
|
4OF5
 
 | |
4OFV
 
 | |
4P3B
 
 | |
4P3A
 
 | |
4P39
 
 | |
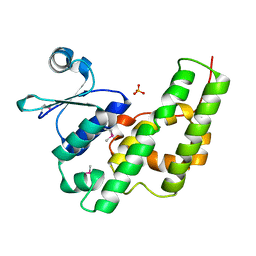
4P2Y
 
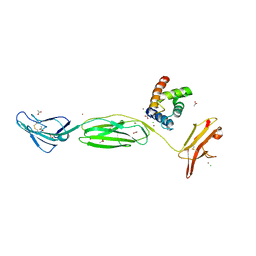 | |
1NHY
 
 | | Crystal Structure of the GST-like Domain of Elongation Factor 1-gamma from Saccharomyces cerevisiae. | | Descriptor: | Elongation factor 1-gamma 1, SULFATE ION | | Authors: | Jeppesen, M.G, Ortiz, P, Kinzy, T.G, Andersen, G.R, Nyborg, J. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Glutathione S-Transferase-like Domain of Elongation Factor 1B{gamma} from Saccharomyces cerevisiae.
J.Biol.Chem., 278, 2003
|
|
1PBU
 
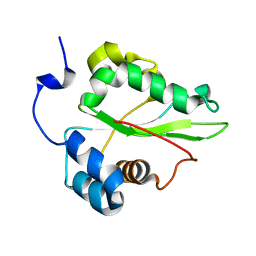 | | Solution structure of the C-terminal domain of the human eEF1Bgamma subunit | | Descriptor: | Elongation factor 1-gamma | | Authors: | Vanwetswinkel, S, Kriek, J, Andersen, G.R, Guntert, P, Dijk, J, Canters, G.W, Siegal, G. | | Deposit date: | 2003-05-15 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | 1H, (15)N and (13)C resonance assignments of the highly conserved 19 kDa C-terminal domain from human Elongation Factor 1Bgamma.
J.Biomol.Nmr, 26, 2003
|
|
1PAQ
 
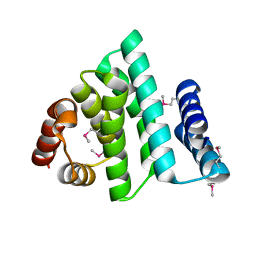 | |
