8TLE
 
 | |
8TLF
 
 | | CDCA7 (Mouse) Binds Non-B-form DNA oligo 36-mer (sg C2-Form 2) | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, GLYCEROL, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8TLG
 
 | | CDCA7 (Mouse) Binds Non-B-form 34-mer DNA oligo | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (34-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
8TLH
 
 | | CDCA7 (Mouse) Binds Non-B-form 32-mer DNA oligo | | Descriptor: | 1,2-ETHANEDIOL, Cell division cycle-associated protein 7, DNA (32-MER), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-21 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Biorxiv, 2023
|
|
6KSQ
 
 | | Middle Domain of Human HSP90 Alpha | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Su, H.X, Zhou, C, Zhang, N.X, Xu, Y.C. | | Deposit date: | 2019-08-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Allosteric Regulation of Hsp90 alpha's Activity by Small Molecules Targeting the Middle Domain of the Chaperone.
Iscience, 23, 2020
|
|
6A71
 
 | | Crystal Structure of Human ATP7B and TM Complex | | Descriptor: | ATP7B protein, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chen, W.B. | | Deposit date: | 2018-06-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tetrathiomolybdate induces dimerization of the metal-binding domain of ATPase and inhibits platination of the protein.
Nat Commun, 10, 2019
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
7X6Z
 
 | |
7X70
 
 | |
7X6Y
 
 | |
8KI5
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
8KIH
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
7E1F
 
 | | Native-DBD | | Descriptor: | DNA-binding response regulator | | Authors: | Hong, S, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
7E1D
 
 | | Se-DBD | | Descriptor: | DNA-binding response regulator | | Authors: | Hong, S, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
6B1E
 
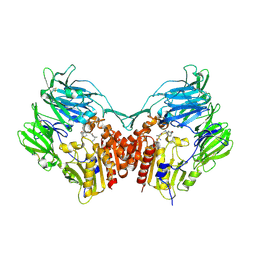 | | The structure of DPP4 in complex with Vildagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(1r,3s,5R,7S)-3-hydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]amino}-1-{(2S)-2-[(E)-iminomethyl]pyrrolidin-1-yl}ethan-1-o ne, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
6B1O
 
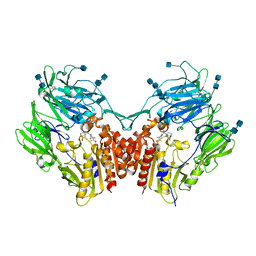 | | The structure of DPP4 in complex with Vildagliptin Analog | | Descriptor: | (2S)-2-amino-1-[(1S,3S,5S)-3-(aminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(1r,3R,5S,7S)-3,5-dihydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
8K3C
 
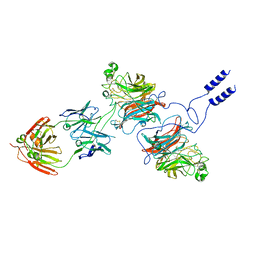 | | Nipah virus Attachment glycoprotein with 41-6 antibody fragment | | Descriptor: | Glycoprotein G, Heavy chain of 41-6 Fab fragments, Light chain of 41-6 Fab fragment | | Authors: | Sun, M.M. | | Deposit date: | 2023-07-15 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Potent human neutralizing antibodies against Nipah virus derived from two ancestral antibody heavy chains.
Nat Commun, 15, 2024
|
|
6BEK
 
 | |
7ECY
 
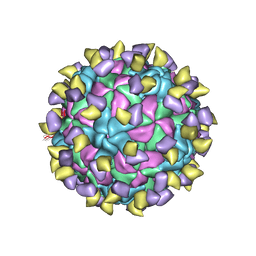 | | EV-D68 in complex with 2H12 Fab (State 3) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EBZ
 
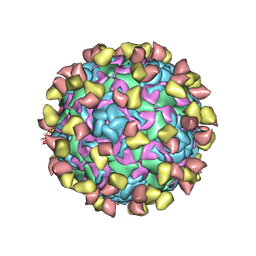 | | EV-D68 in complex with 2H12 Fab (state S1) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EBR
 
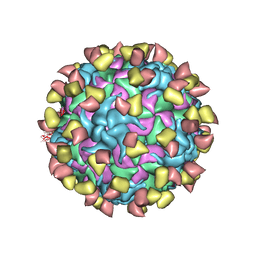 | | EV-D68 in complex with 2H12 Fab (state S2) | | Descriptor: | 2H12 Fab heavy chain, 2H12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EC5
 
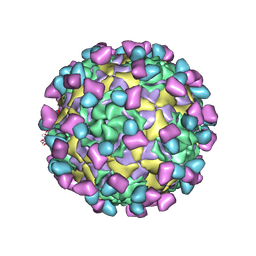 | | EV-D68 in complex with 8F12 Fab | | Descriptor: | 8F12 Fab heavy chain, 8F12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
4N0N
 
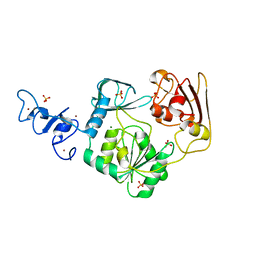 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
6LWA
 
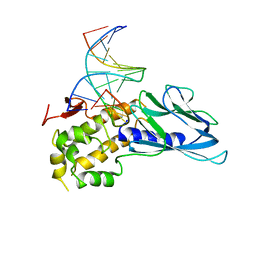 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
