8ZNZ
 
 | | CD73 bound with HB0045 | | Descriptor: | 5'-nucleotidase, CALCIUM ION, HB0038 Fab heavy chain, ... | | Authors: | Xu, J, Wan, L, He, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | An antibody cocktail targeting two different CD73 epitopes enhances enzyme inhibition and tumor control.
Nat Commun, 15, 2024
|
|
6W6G
 
 | |
6W6H
 
 | |
6W6J
 
 | |
6W6I
 
 | |
6W6E
 
 | |
6XUD
 
 | | Apo Ab 1116NS19.9 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUL
 
 | | Apo Ab 5b1 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
6VLB
 
 | |
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6PYH
 
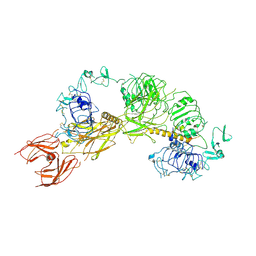 | | Cryo-EM structure of full-length IGF1R-IGF1 complex. Only the extracellular region of the complex is resolved. | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Choi, E, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of the activation of type 1 insulin-like growth factor receptor.
Nat Commun, 10, 2019
|
|
6XUK
 
 | | AbLIFT design 15 of Ab 1116NS19.9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6PXW
 
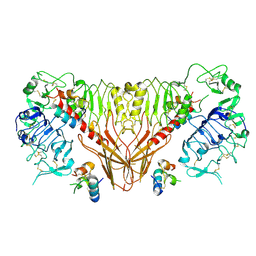 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6VLC
 
 | |
6XTG
 
 | | Ab 1116NS19.9 bound to CA19-9 | | Descriptor: | Heavy chain, Light chain, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6WML
 
 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
6XUN
 
 | | Ab 5b1 bound to CA19-9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
4QPM
 
 | | Structure of Bub1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lin, Z.H, Jia, L.Y, Tomchick, D.R, Luo, X.L, Yu, H.T. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Substrate-Specific Activation of the Mitotic Kinase Bub1 through Intramolecular Autophosphorylation and Kinetochore Targeting.
Structure, 22, 2014
|
|
7XE4
 
 | | structure of a membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
1QWF
 
 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1QWE
 
 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | Descriptor: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7END
 
 | |
