7WL9
 
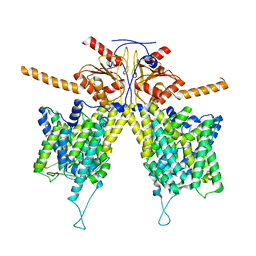 | | Mouse Pendrin in chloride and bicarbonate in asymmetric state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-12 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WLA
 
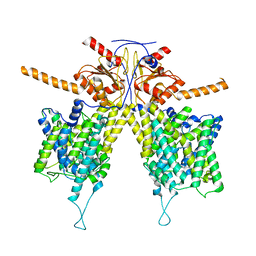 | |
7WK7
 
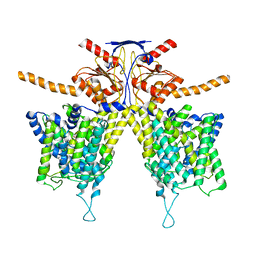 | | Mouse Pendrin bound bicarbonate in inward state | | Descriptor: | BICARBONATE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
5BPY
 
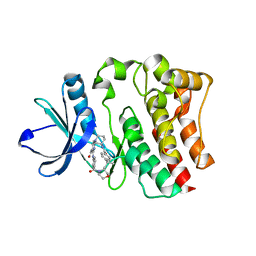 | |
7WL8
 
 | |
7WLB
 
 | |
5BQ0
 
 | |
6NNJ
 
 | |
6NNF
 
 | |
6NM6
 
 | |
5U1F
 
 | | Initial contact of HIV-1 Env with CD4: Cryo-EM structure of BG505 DS-SOSIP trimer in complex with CD4 and antibody PGT145 | | Descriptor: | BG505 DS-SOSIP gp120, BG505 SOSIP gp41, PGT145 heavy chain, ... | | Authors: | Acharya, P, Kwong, P.D, Potter, C.S, Carragher, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-02-22 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7EZK
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gs complex | | Descriptor: | Chimera of Guanine nucleotide-binding protein G(i) subunit alpha-1 and Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZM
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZH
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
2NN6
 
 | | Structure of the human RNA exosome composed of Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, Rrp42, Csl4, Rrp4, and Rrp40 | | Descriptor: | 3'-5' exoribonuclease CSL4 homolog, Exosome complex exonuclease RRP4, Exosome complex exonuclease RRP40, ... | | Authors: | Lima, C.D. | | Deposit date: | 2006-10-23 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Reconstitution, activities, and structure of the eukaryotic RNA exosome.
Cell(Cambridge,Mass.), 127, 2006
|
|
5YE9
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(3-cyano-4-naphthalen-2-yloxy-phenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YE7
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(4-naphthalen-2-yloxyphenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YE8
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[3,4-bis(fluoranyl)phenyl]methanesulfonamide, Platelet-activating factor acetylhydrolase | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YEA
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | 4-[[4-[4-chloranyl-3-(trifluoromethyl)phenoxy]-3-cyano-phenyl]sulfamoyl]benzoic acid, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8XK2
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | Spike protein S1, VHH60 nanobody | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8XKI
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
