2HQ6
 
 | | Structure of the Cyclophilin_CeCYP16-Like Domain of the Serologically Defined Colon Cancer Antigen 10 from Homo Sapiens | | Descriptor: | GLYCEROL, IODIDE ION, Serologically defined colon cancer antigen 10 | | Authors: | Walker, J.R, Davis, T, Paramanathan, R, Newman, E.M, Finerty Jr, P.J, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
1VLR
 
 | |
1VKM
 
 | |
1VQR
 
 | |
1VPZ
 
 | |
1VKH
 
 | |
4D9S
 
 | | Crystal structure of Arabidopsis thaliana UVR8 (UV Resistance locus 8) | | Descriptor: | UVB-resistance protein UVR8 | | Authors: | Arvai, A.S, Christie, J.M, Pratt, A.J, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges.
Science, 335, 2012
|
|
3IPR
 
 | |
1ZX8
 
 | |
1Z9F
 
 | |
3GVA
 
 | |
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
6W8W
 
 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
6W8V
 
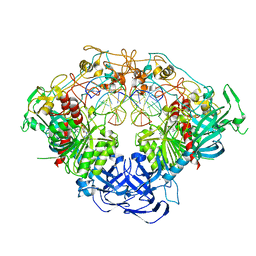 | | Crystal structure of mouse DNMT1 in complex with ACG DNA | | Descriptor: | ACG DNA (5'-D(*AP*CP*TP*TP*AP*(C49)P*GP*GP*AP*AP*GP*G)-3'), ACG DNA (5'-D(*CP*CP*TP*TP*CP*(5CM)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
4HDU
 
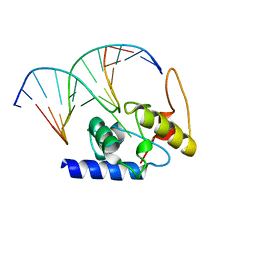 | |
6EL3
 
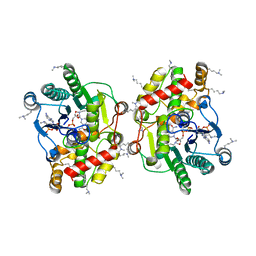 | | Structure of Progesterone 5beta-Reductase from Arabidopsis thaliana in complex with NADP | | Descriptor: | 3-oxo-Delta(4,5)-steroid 5-beta-reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Muller, Y.A, Schmidt, K, Egerer-Sieber, C. | | Deposit date: | 2017-09-27 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | PRISEs (progesterone 5 beta-reductase and/or iridoid synthase-like 1,4-enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism.
Phytochemistry, 156, 2018
|
|
4HDV
 
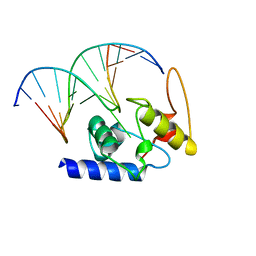 | |
2I8C
 
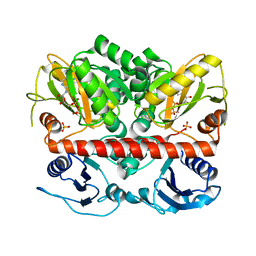 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I87
 
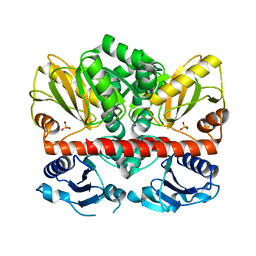 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | D-alanine-D-alanine ligase, SULFATE ION | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4L1F
 
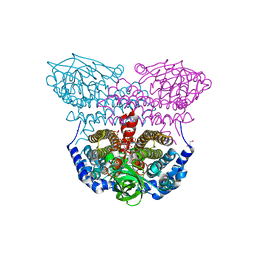 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | 1,3-PROPANDIOL, Acyl-CoA dehydrogenase domain protein, COENZYME A PERSULFIDE, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L2I
 
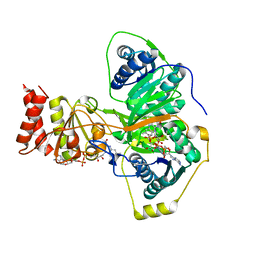 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4KPU
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4ENJ
 
 | |
4ENM
 
 | |
4ENK
 
 | |
