8SIS
 
 | |
8SIR
 
 | |
8SIQ
 
 | |
7RWE
 
 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7RXO
 
 | | Crystal structure of CDK2 liganded with compound WN333 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(methoxycarbonyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S4T
 
 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S7A
 
 | | Crystal structure of CDK2 liganded with compound EF3019 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(2H-indazol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S85
 
 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
4TR7
 
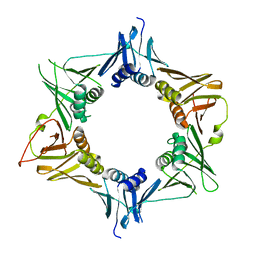 | |
4TR8
 
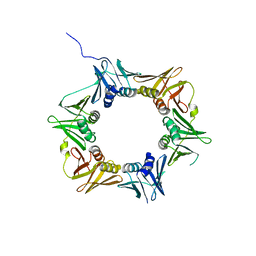 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TSZ
 
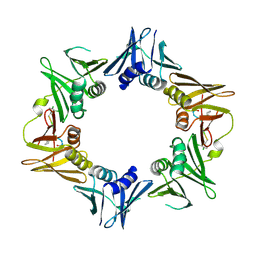 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
8FG1
 
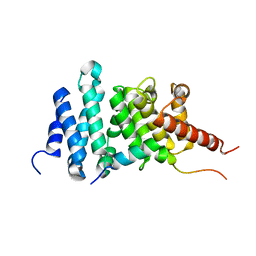 | | Human diaphanous inhibitory domain bound to diaphanous autoregulatory domain | | Descriptor: | Protein diaphanous homolog 1 | | Authors: | Ramirez, L.M.S, Theophall, G, Premo, A, Manigrasso, M, Yepuri, G, Burz, D, Ramasamy, R, Schmidt, A.M, Shekhtman, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of the productive encounter complex results in dysregulation of DIAPH1 activity.
J.Biol.Chem., 299, 2023
|
|
6VAP
 
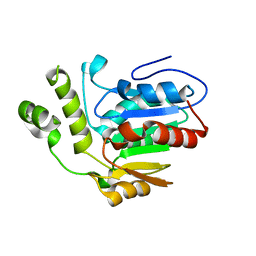 | | Structure of the type II thioesterase BorB from the borrelidin biosynthetic cluster | | Descriptor: | Thioesterase | | Authors: | Pereira, J.H, Curran, S.C, Baluyot, M.-J, Lake, J, Putz, H, Rosenburg, D, Keasling, J, Adams, P.D. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and Function of BorB, the Type II Thioesterase from the Borrelidin Biosynthetic Gene Cluster.
Biochemistry, 59, 2020
|
|
7KZB
 
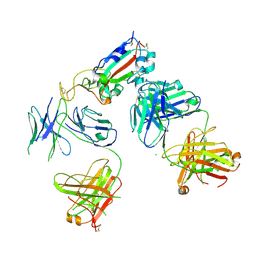 | |
7KZC
 
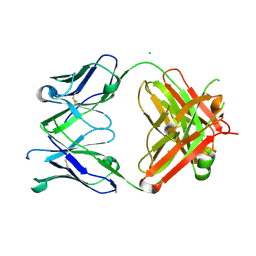 | |
7KZA
 
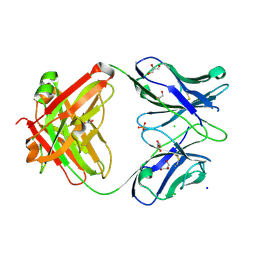 | |
4BEL
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-11 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BFB
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, XA4813 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKQ
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZL7
 
 | | BACE2 FYNOMER COMPLEX | | Descriptor: | BETA-SECRETASE 2, FYNOMER 2B-H11 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKG
 
 | | BACE2 MUTANT APO STRUCTURE | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7BDK
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDL
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ADP | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDJ
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ATPgammaS | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDI
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
