4TTY
 
 | |
4U2I
 
 | |
5D5P
 
 | | HcgB from Methanococcus maripaludis | | Descriptor: | HcgB | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
8OKB
 
 | | SARS-CoV2 NSP5 in complex with a peptidomimetic ligand | | Descriptor: | 3C-like proteinase nsp5, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
8OKC
 
 | | SARS-CoV2 NSP5 in complex with a GC-376 based peptidomimetic PROTAC | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-1-[[(~{Z},2~{S})-5-[4-[[1-[2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]-6-fluoranyl-1,3-bis(oxidanylidene)isoindol-5-yl]piperidin-4-yl]methyl]piperazin-1-yl]-5-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]pent-3-en-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
6JEO
 
 | | Structure of PSI tetramer from Anabaena | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
4D7W
 
 | | Crystal structure of sortase C1 (SrtC1) from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, SORTASE FAMILY PROTEIN | | Authors: | Malito, E, Lazzarin, M, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-11-28 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Noncanonical Sortase-Mediated Assembly of Pilus Type 2B in Group B Streptococcus.
Faseb J., 29, 2015
|
|
7VUH
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUG
 
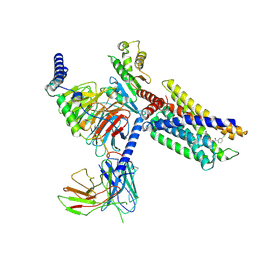 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUJ
 
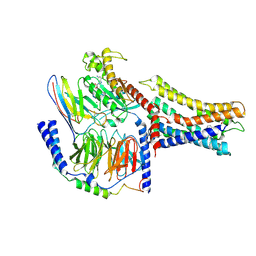 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUI
 
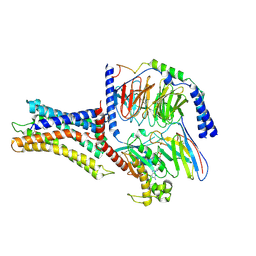 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VD6
 
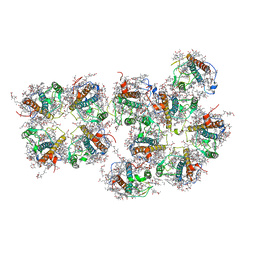 | | Structure of S1M1-type FCPII complex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
7VD5
 
 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
7PKI
 
 | |
6ZUC
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1-Lig1-HAB1 TERNARY COMPLEX | | Descriptor: | 1,4-dimethyl-2-oxidanylidene-~{N}-(phenylmethyl)quinoline-6-sulfonamide, CHLORIDE ION, CSPYL1, ... | | Authors: | Albert, A, Infantes, L, Benavente, J.L. | | Deposit date: | 2020-07-22 | | Release date: | 2021-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
5TWN
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA- DEPENDENT RNA POLYMERASE IN COMPLEX WITH 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 5-[3-(tert-butylcarbamoyl)phenyl]-6-(ethylamino)-2-(4-fluorophenyl)-N-methylfuro[2,3-b]pyridine-3-carboxamide, NS5B RNA- DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The discovery of a pan-genotypic, primer grip inhibitor of HCV NS5B polymerase.
Medchemcomm, 8, 2017
|
|
4DS7
 
 | |
7F4V
 
 | | Cryo-EM structure of a primordial cyanobacterial photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nagao, R, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-06-21 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural basis for the absence of low-energy chlorophylls responsible for photoprotection from a primitive cyanobacterial PSI
Biorxiv, 2022
|
|
7Q8I
 
 | | Peptide AVAEKQ in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AVAEKQ peptide, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8O
 
 | | Peptide LLSGKE in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8H
 
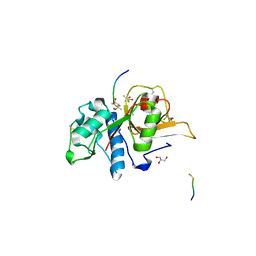 | | Peptide EVCKKKK in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q9H
 
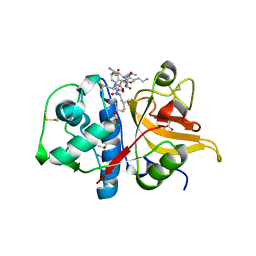 | | Peptide LLKAVAEKQ in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8D
 
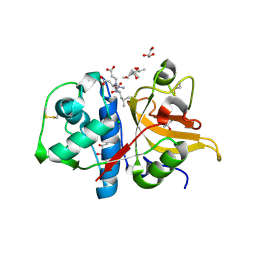 | | Peptide TRESEDLE in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ASP-LEU-GLU(AMI), CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8G
 
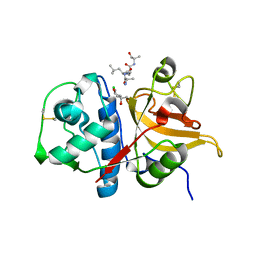 | | Peptide ALAASS in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALAASS Peptide, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7Q8J
 
 | | Peptide IILKEK in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cathepsin L2, EYS Peptide, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
