4HFZ
 
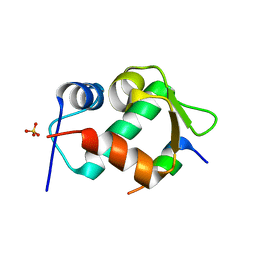 | | Crystal Structure of an MDM2/P53 Peptide Complex | | Descriptor: | Cellular tumor antigen p53, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Anil, B, Riedinger, C, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HIZ
 
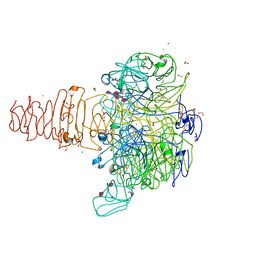 | | Phage phi92 endosialidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schwarzer, D, Browning, C, Leiman, P.G. | | Deposit date: | 2012-10-12 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of Endosialidase from Phage phi92 that cleaves alpha2,8- and alpha2,9-linked polysialic acid
To be Published
|
|
6XUC
 
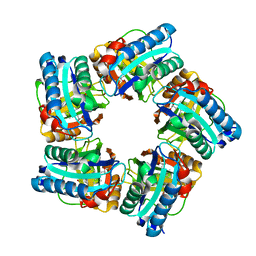 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6XIW
 
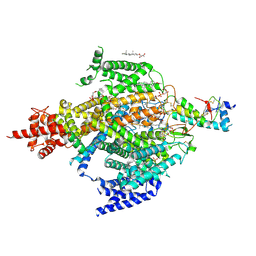 | | Cryo-EM structure of the sodium leak channel NALCN-FAM155A complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Noland, C.L, Weidling, C, Clairfeuille, T, Bahlke, O.O, Ameen, A.O, Li, Z.R, Arthur, C.P, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human sodium leak channel NALCN.
Nature, 587, 2020
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
8R3E
 
 | | Huntingtin, 1-17, MBP-N | | Descriptor: | Maltodextrin-binding protein, ZINC ION | | Authors: | Steinbacher, S, Toledo-Sherman, L, Dominguez, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Post-translational modifications in the first 17 amino acids of huntingtin influence self-association and interaction with membranes
To Be Published
|
|
7ATI
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7ASB
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
6VWM
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
7ASX
 
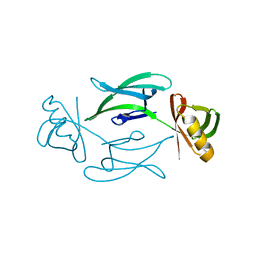 | | Fixed-target serial femtosecond crystallography using in cellulo grown Neurospora crassa HEX-1 microcrystals. (Chip 1) | | Descriptor: | eIF-5a domain-containing protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Barthelmess, M, Fischer, P, Seuring, C, Wagner, A, Meents, A, Redecke, L. | | Deposit date: | 2020-10-28 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals.
Iucrj, 8, 2021
|
|
7ASI
 
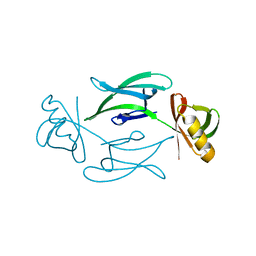 | | Fixed-target serial femtosecond crystallography using in cellulo grown Neurospora crassa HEX-1 microcrystals. (Chips 1+2) | | Descriptor: | eIF-5a domain-containing protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Barthelmess, M, Fischer, P, Seuring, C, Wagner, A, Meents, A, Redecke, L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals.
Iucrj, 8, 2021
|
|
8BTM
 
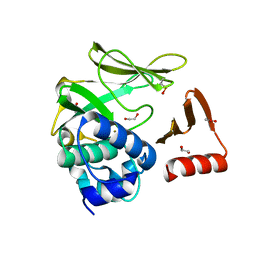 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
8OGI
 
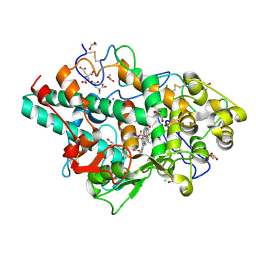 | | Structure of native human eosinophil peroxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pfanzagl, V, Obinger, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Posttranslational modification and heme cavity architecture of human eosinophil peroxidase-insights from first crystal structure and biochemical characterization.
J.Biol.Chem., 299, 2023
|
|
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
8OWD
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L1 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWJ
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWE
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8Q1O
 
 | | S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 32-209), important for Self-assembly | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Sagmeister, T, Grininger, C, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
8B69
 
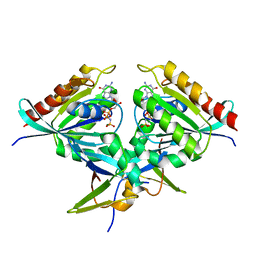 | | Heterotetramer of K-Ras4B(G12V) and Rgl2(RBD) | | Descriptor: | Isoform 2B of GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tariq, M, Fairall, L, Romartinez-Alonso, B, Dominguez, C, Schwabe, J.W.R, Tanaka, K. | | Deposit date: | 2022-09-26 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
6VWN
 
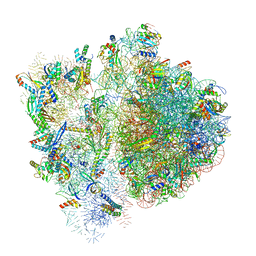 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
8OIU
 
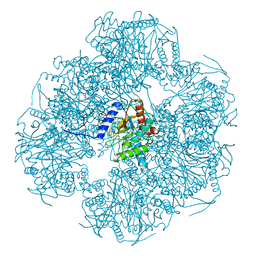 | | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Skalidis, I, Tueting, C, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon.
Commun Biol, 6, 2023
|
|
