3C3B
 
 | | Crystal Structure of human phosphoglycerate kinase bound to D-CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, Phosphoglycerate kinase 1 | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
3BX7
 
 | |
3C3A
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
1H6O
 
 | | Dimerisation domain from human TRF1 | | Descriptor: | TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Fairall, L, Chapman, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2.
Mol.Cell, 8, 2001
|
|
1NHV
 
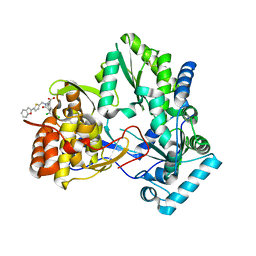 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NHU
 
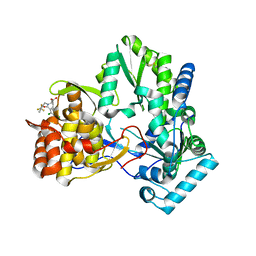 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1E8C
 
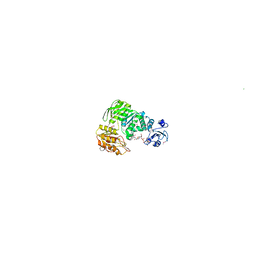 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1E0D
 
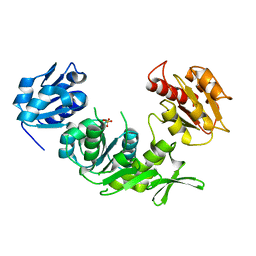 | | UDP-N-Acetylmuramoyl-L-Alanine:D-Glutamate Ligase | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Fanchon, E, Bertrand, J, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-03-24 | | Release date: | 2000-06-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Open" Structures of Murd: Domain Movements and Structural Similarities with Folylpolyglutamate Synthetase.
J.Mol.Biol., 301, 2000
|
|
1MQO
 
 | |
4AA6
 
 | | The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation | | Descriptor: | 5'-D(*CP*TP*AP*AP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP *CP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP *CP*TP*TP*A)-3', ESTROGEN RECEPTOR, ... | | Authors: | Schwabe, J.W, Chapman, L, Rhodes, D. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation.
Structure, 3, 1995
|
|
4ACJ
 
 | | Crystal structure of the TLDC domain of Oxidation resistance protein 2 from zebrafish | | Descriptor: | WU:FB25H12 PROTEIN, | | Authors: | Blaise, M, B Alsarraf, H.M.A, Wong, J.E.M.M, Midtgaard, S.R, Laroche, F, Schack, L, Spaink, H, Stougaard, J, Thirup, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of the Tldc Domain of Oxidation Resistance Protein 2 from Zebrafish.
Proteins, 80, 2012
|
|
2GIG
 
 | | Alteration of sequence specificity of the type II restriction endonuclease HINCII through an indirect readout mechanism | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*GP*C)-3', SODIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
2GIE
 
 | | HincII bound to cognate DNA GTTAAC | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*TP*AP*AP*CP*CP*GP*G)-3', SODIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
1HCQ
 
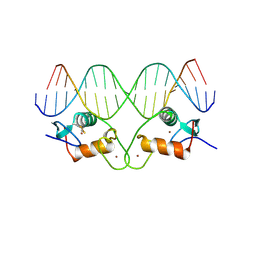 | | THE CRYSTAL STRUCTURE OF THE ESTROGEN RECEPTOR DNA-BINDING DOMAIN BOUND TO DNA: HOW RECEPTORS DISCRIMINATE BETWEEN THEIR RESPONSE ELEMENTS | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP*CP*CP*T P*G)-3'), DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP*CP*CP*T P*G)-3'), PROTEIN (ESTROGEN RECEPTOR), ... | | Authors: | Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1995-01-04 | | Release date: | 1995-11-23 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements.
Cell(Cambridge,Mass.), 75, 1993
|
|
2GIH
 
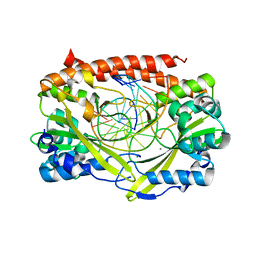 | | Q138F HincII bound to cognate DNA GTCGAC and Ca2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*GP*C)-3', CALCIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
3C3C
 
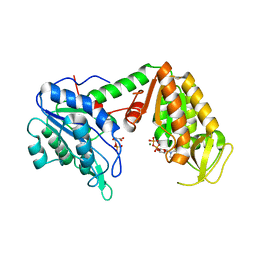 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate and L-CDP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
3BX8
 
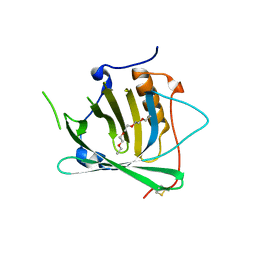 | | Engineered Human Lipocalin 2 (LCN2), apo-form | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ENGINEERED HUMAN LIPOCALIN 2, PENTAETHYLENE GLYCOL | | Authors: | Schonfeld, D.L, Chatwell, L, Skerra, A. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High affinity molecular recognition and functional blockade of CTLA-4 by an engineered human lipocalin
To be Published
|
|
1DYP
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
1FFL
 
 | |
4N57
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-IVa ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, APH(2'')-Id, MAGNESIUM ION | | Authors: | Kaplan, E, Leban, N, Chaloin, L, Guichou, J.-F, Lionne, C. | | Deposit date: | 2013-10-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-IVa ADP complex
To be Published
|
|
2K1Z
 
 | |
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1FWM
 
 | | Crystal structure of the thymidylate synthase R166Q mutant | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Sotelo-Mundo, R.R, Changchien, L, Maley, F, Montfort, W.R. | | Deposit date: | 2000-09-23 | | Release date: | 2003-11-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thymidylate synthase mutant R166Q: Structural basis for the nearly complete loss of catalytic activity.
J.Biochem.Mol.Toxicol., 20, 2006
|
|
