1QK9
 
 | | The solution structure of the domain from MeCP2 that binds to methylated DNA | | Descriptor: | METHYL-CPG-BINDING PROTEIN 2 | | Authors: | Wakefield, R.I.D, Smith, B.O, Nan, X, Free, A, Soteriou, A, Uhrin, D, Bird, A.P, Barlow, P.N. | | Deposit date: | 1999-07-12 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Domain from Mecp2 that Binds to Methylated DNA
J.Mol.Biol., 291, 1999
|
|
3BX7
 
 | |
4MI0
 
 | | Human Enhancer of Zeste (Drosophila) Homolog 2(EZH2) | | Descriptor: | Histone-lysine N-methyltransferase EZH2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations.
Plos One, 8, 2013
|
|
3C73
 
 | | Structure of CEHC variant ResA | | Descriptor: | SULFATE ION, Thiol-disulfide oxidoreductase resA | | Authors: | Crow, A. | | Deposit date: | 2008-02-06 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effects of substitutions in the CXXC active-site motif of the extracytoplasmic thioredoxin ResA.
Biochem.J., 414, 2008
|
|
3BLJ
 
 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ADP-ribose) polymerase 15, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
6W1P
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.26 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3BK7
 
 | |
6W1R
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2KKY
 
 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
3C3E
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and GDP. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
1QGI
 
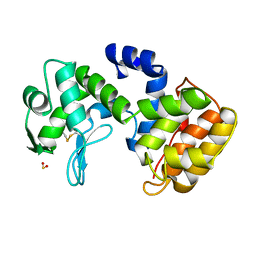 | | CHITOSANASE FROM BACILLUS CIRCULANS | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | Authors: | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
8HH8
 
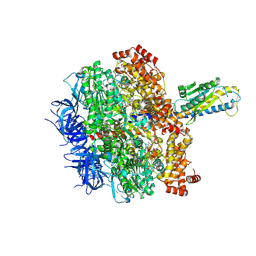 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH6
 
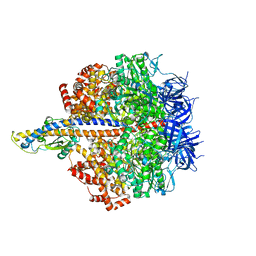 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH7
 
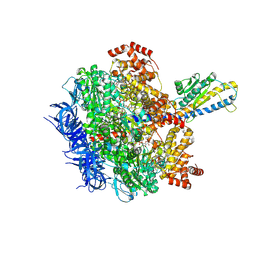 | | F1 domain of FoF1-ATPase from Bacillus PS3, 81 degrees, lowATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH5
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,highATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HUK
 
 | | X-ray structure of human PPAR alpha ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUL
 
 | | X-ray structure of human PPAR delta ligand binding domain-lanifibranor co-crystals obtained by co-crystallization | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUQ
 
 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUN
 
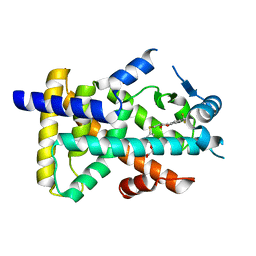 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUP
 
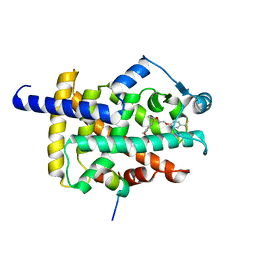 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUM
 
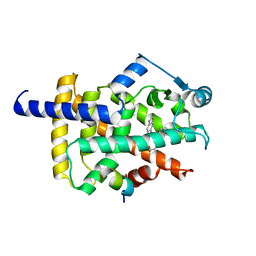 | | X-ray structure of human PPAR gamma ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
6W1V
 
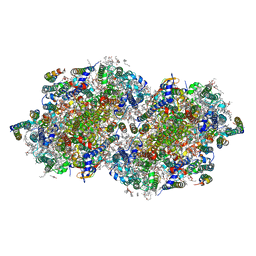 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HUO
 
 | | X-ray structure of human PPAR delta ligand binding domain-seladelpar co-crystals obtained by co-crystallization | | Descriptor: | Peroxisome proliferator-activated receptor delta, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HHV
 
 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
6W1O
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
