4WU2
 
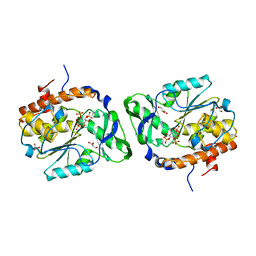 | | Structure of the PTP-like myo-inositol phosphatase from Selenomonas ruminantium in complex with myo-inositol-(1,4,5)-trikisphosphate | | Descriptor: | CHLORIDE ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,4,5)-trikisphosphate
To Be Published
|
|
4X9C
 
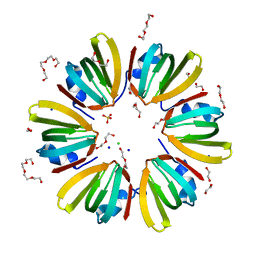 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
4GRB
 
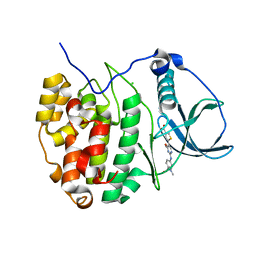 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
1ADU
 
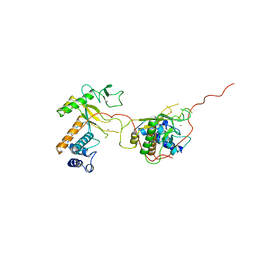 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
4YPD
 
 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
4J6X
 
 | |
1AXG
 
 | | CRYSTAL STRUCTURE OF THE VAL203->ALA MUTANT OF LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH COFACTOR NAD AND INHIBITOR TRIFLUOROETHANOL SOLVED TO 2.5 ANGSTROM RESOLUTION | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Bahnson, B.J, Goldstein, B.M, Klinman, J.P. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1ADB
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLNICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
1AJ6
 
 | |
1BBU
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) COMPLEXED WITH LYSINE | | Descriptor: | LYSINE, PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
1BBW
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) | | Descriptor: | PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
5ADH
 
 | |
4J6Y
 
 | |
1AH0
 
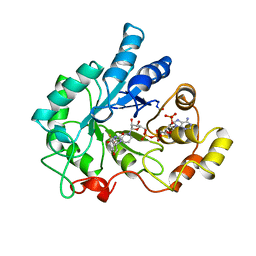 | | PIG ALDOSE REDUCTASE COMPLEXED WITH SORBINIL | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SORBINIL | | Authors: | Moras, D, Podjarny, A.D. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
1ADV
 
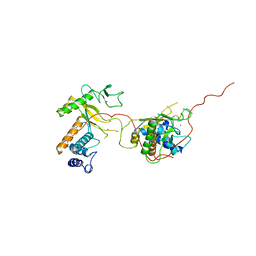 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1AH4
 
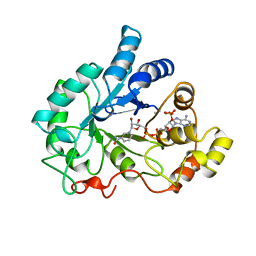 | | PIG ALDOSE REDUCTASE, HOLO FORM | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Moras, D, Podjarny, A. | | Deposit date: | 1997-04-12 | | Release date: | 1998-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
4J6W
 
 | | Crystal structure of HFQ from Pseudomonas aeruginosa in complex with CTP | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Nikulin, A.D, Murina, V, Lekontseva, N. | | Deposit date: | 2013-02-12 | | Release date: | 2013-07-31 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ADC
 
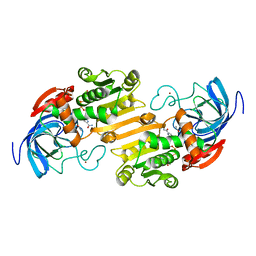 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
1BYL
 
 | |
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
2J34
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1-BENZOTHIOPHENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Senger, S, Convery, M.A, Chan, C, Watson, N.S. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Arylsulfonamides: A Study of the Relationship between Activity and Conformational Preferences for a Series of Factor Xa Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2J38
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1-BENZOTHIOPHENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Senger, S, Convery, M.A, Chan, C, Watson, N.S. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arylsulfonamides: A Study of the Relationship between Activity and Conformational Preferences for a Series of Factor Xa Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3QT6
 
 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor DPGP | | Descriptor: | 1-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}acetyl)-L-proline, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3QSY
 
 | | Recognition of the methionylated initiator tRNA by the translation initiation factor 2 in Archaea | | Descriptor: | METHIONINE, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Translation initiation factor 2 subunit alpha, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Zelinskaya, N.V, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2011-02-22 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the archaeal translation initiation factor 2 in complex with a GTP analogue and Met-tRNAf(Met.)
J.Mol.Biol., 425, 2013
|
|
3QT7
 
 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
