7RA3
 
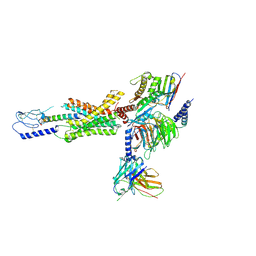 | | cryo-EM of human Gastric inhibitory polypeptide receptor GIPR bound to GIP | | Descriptor: | Gastric inhibitory polypeptide, Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural determinants of dual incretin receptor agonism by tirzepatide.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RPR
 
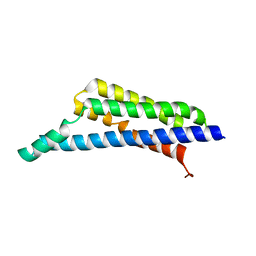 | | Borrelia miyamotoi FbpA complement inhibitory domain | | Descriptor: | Fibronectin-binding lipoprotein FbpA | | Authors: | Booth, C.E, Garcia, B.L. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Borrelia miyamotoi FbpA and FbpB Are Immunomodulatory Outer Surface Lipoproteins With Distinct Structures and Functions.
Front Immunol, 13, 2022
|
|
7RPS
 
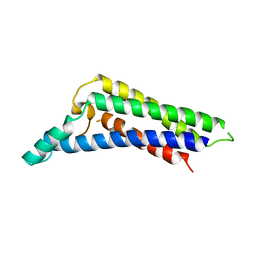 | | B. miyamotoi FbpB complement inhibitory domain | | Descriptor: | Fibronectin-binding lipoprotein FbpB | | Authors: | Booth, C.E, Garcia, B.L. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Borrelia miyamotoi FbpA and FbpB Are Immunomodulatory Outer Surface Lipoproteins With Distinct Structures and Functions.
Front Immunol, 13, 2022
|
|
6NDL
 
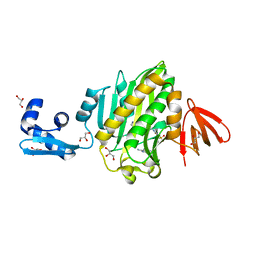 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
6NKH
 
 | |
7POK
 
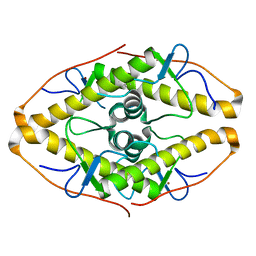 | | Crystal structure of ZAD-domain of Pita protein from D.melanogaster | | Descriptor: | LD15650p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7PPP
 
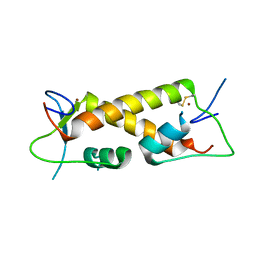 | | Crystal structure of ZAD-domain of ZNF_276 protein from rabbit. | | Descriptor: | ZINC ION, Zinc finger protein 276 | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7POH
 
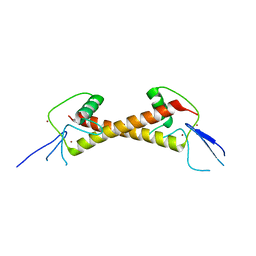 | | Crystal structure of ZAD-domain of Serendipity-d protein from D.melanogaster | | Descriptor: | Serendipity locus protein delta, ZINC ION | | Authors: | Boyko, K.M, Kachalova, G.S, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7PO9
 
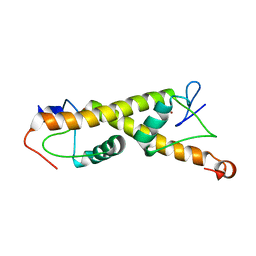 | | Crystal structure of ZAD-domain of M1BP protein from D.melanogaster | | Descriptor: | LD30467p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
6NXE
 
 | |
7R6J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 1 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[3-methyl-5-(pyrimidin-2-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7R8P
 
 | |
7R8Q
 
 | | Closed form of SAOUHSC_02373 in complex with ADP, citrate, Mg2+ and Na+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, CITRIC ACID, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-06-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of an ʟ-amino acid ligase implicated in Staphylococcal sulfur amino acid metabolism.
J.Biol.Chem., 298, 2022
|
|
7R1E
 
 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7REV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7ROF
 
 | |
7RNQ
 
 | |
7RNP
 
 | |
6OWD
 
 | |
6OWU
 
 | |
7RD2
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 2 | | Descriptor: | (2R,3R,4R,5S)-1-{[4-({4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-2-nitroanilino}methyl)phenyl]methyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-09 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6OPM
 
 | | Casposase bound to integration product | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, DNA 21-mer, ... | | Authors: | Dyda, F, Hickman, A.B, Kailasan, S. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Elife, 9, 2020
|
|
6O9B
 
 | |
6OKD
 
 | | Crystal Structure of human transferrin receptor in complex with a cystine-dense peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A TfR-Binding Cystine-Dense Peptide Promotes Blood-Brain Barrier Penetration of Bioactive Molecules.
J.Mol.Biol., 432, 2020
|
|
6ORU
 
 | |
